5. LabelTransfer full unintegrated¶
suppressPackageStartupMessages({
library(reticulate)
library(Seurat)
library(dplyr)
library(ggpubr)
library(sceasy)
library(future)
})
# utilities
o<- function(w,h) options(repr.plot.width=w, repr.plot.height=h)
manycolors=c('#e6194b', '#3cb44b', '#ffe119', '#4363d8', '#f58231',
'#911eb4', '#46f0f0', '#f032e6', '#bcf60c', '#fabebe',
'#008080', '#e6beff', '#9a6324', '#fffac8', '#800000',
'#aaffc3', '#808000', '#ffd8b1', '#000075', '#808080',
'#4f34ff', '#f340F0')
covid.color.table= c(
'B'="#FFF3B0", 'Plasma'='#E09F3E',
'T'='#2F6D9D',
'T CD4'='#68BDDF',
'T CD8'='#FF046E',
'CD8 cytotoxic T'='#9D2A2B',
'T cycling'='#ADA7FF',
'Treg'='#493A9D',
'NKT'='#3300FF',
'NK'='#D543FF',
'gdT'='#6A24FF',
'Neu'='#312697',
'macrophage'='#429783',
'Mast'='#FF6B3E',
'cDC1'='#FF8F39',
'cDC2'='#E7FF3F',
'moDC'='#FFBCE1',
'pDC'='#FF72DB',
'DC'='#FF0022',
'Mye'='#19C3BE'
)
heca.color.table=c(
'B cell'='#FFF3B0',
'CD8 T cell'='#FF046E',
'Dendritic cell'='#FF0022',
'Macrophage'='#429783',
'Mast cell'='#FF6B3E',
'Megakaryocyte'='gray',
'Monocyte'='#429753',
'Myeloid cell'='#19C3BE',
'NK cell'='#D543FF',
'Neutrophilic granunocyte'='#312697',
'Plasma B cell'='#E09F3E',
'T cell'='#2F6D9D'
)
load data¶
load hECA data¶
# convert h5ad to rds format so that R can read it
sceasy::convertFormat('./lungimm_heca.h5ad', from="anndata", to="seurat",
outFile='lungimm_heca.rds')
X -> counts
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from DMAPharmony_bydonor_ to DMAPharmonybydonor_”
Warning message:
“All keys should be one or more alphanumeric characters followed by an underscore '_', setting key to DMAPharmonybydonor_”
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from PCAharmony_bydonor_ to PCAharmonybydonor_”
Warning message:
“All keys should be one or more alphanumeric characters followed by an underscore '_', setting key to PCAharmonybydonor_”
An object of class Seurat
20492 features across 53299 samples within 1 assay
Active assay: RNA (20492 features, 0 variable features)
6 dimensional reductions calculated: densmap, densmap_harmonyByDonor, dmap_harmony_bydonor, pca, pca_harmony_bydonor, umap
# read saved hECA data
heca = readRDS('./lungimm_heca.rds')
# create a clean seurat object
heca.full = CreateSeuratObject(heca@assays$RNA@data, meta.data = heca@meta.data)
heca.full@assays$RNA@data <- heca.full@assays$RNA@counts
heca.full[["umap"]] <- CreateDimReducObject(heca@reductions$umap@cell.embeddings, key = "umap_")
Warning message:
“No assay specified, setting assay as RNA by default.”
# show different batches
o(15,5)
DimPlot(heca.full, group.by='ann',cols = heca.color.table, split.by = 'study_id', pt.size=0.01, label=T, repel=T)
# print study_id
unique(heca.full@meta.data$study_id)
- 10.1038/s41591-019-0468-5
- 10.1186/s13059-019-1906-x
- 10.1038/s41586-020-2157-4
Levels:
- '10.1038/s41586-020-2157-4'
- '10.1038/s41591-019-0468-5'
- '10.1186/s13059-019-1906-x'
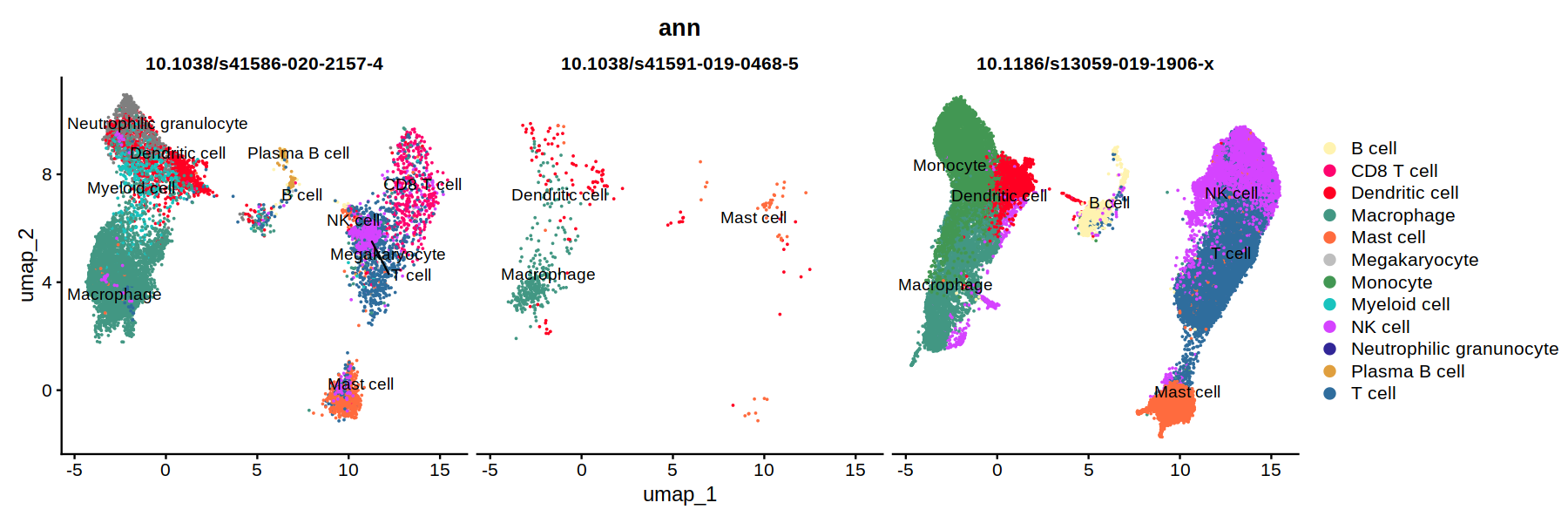
#heca1 = subset(heca.full, study_id=='10.1038/s41586-020-2157-4')
#heca2 = subset(heca.full, study_id=='10.1186/s13059-019-1906-x')
load COVID data¶
load("./nCoV.integrated.annotated.rda")
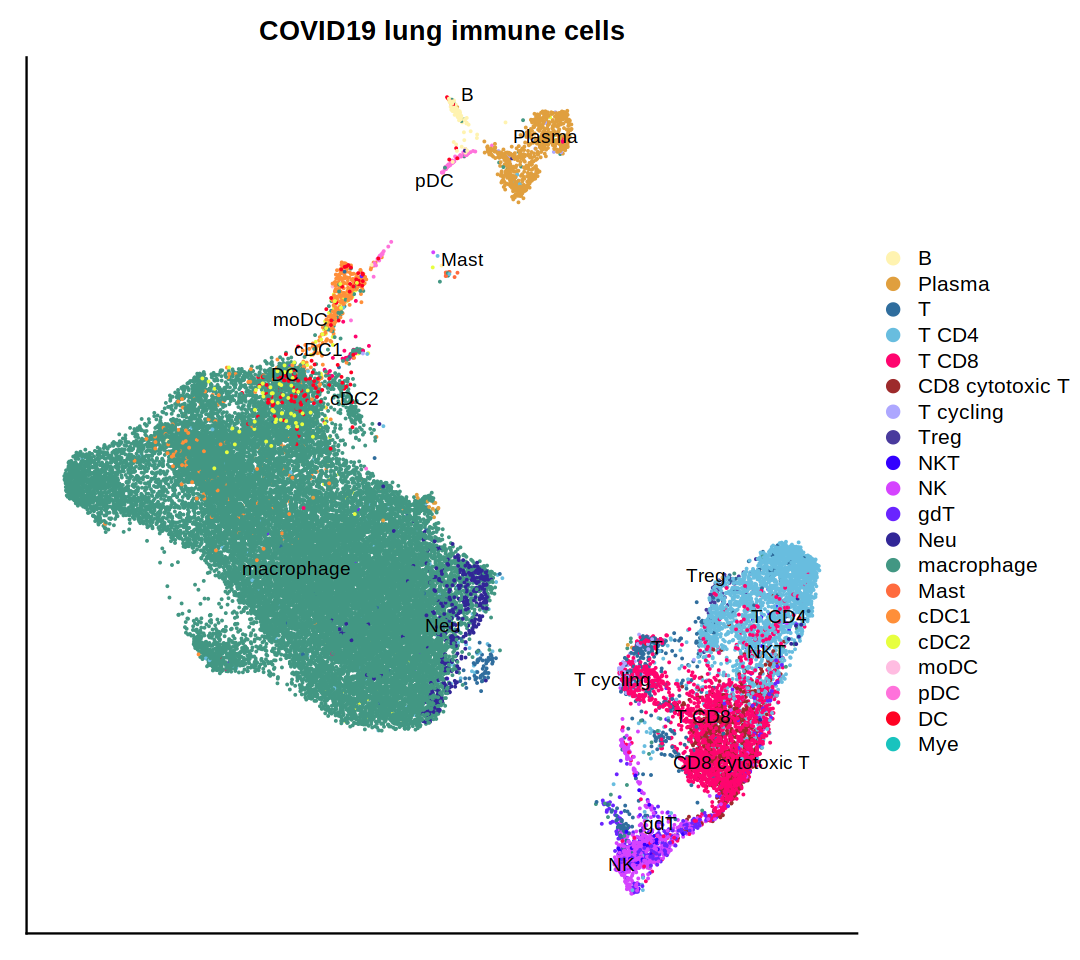
o(9,8)
DimPlot(immune.combined, group.by='persample_ann', label=T, repel=T, cols=covid.color.table) + ggtitle(
"COVID19 lung immune cells")&
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank())
cairo_pdf(filename = 'covid19.umap.pdf', width = 9,height = 8)
DimPlot(immune.combined, group.by='persample_ann', label=T, repel=T, cols=covid.color.table) + ggtitle(
"COVID19 lung immune cells")&
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank())
dev.off()
full batches - unintegrated data transfer¶
# find genes for label transfer
DefaultAssay(heca.full)<-'RNA'
heca.full = FindVariableFeatures(heca.full, selection.method="vst", nfeatures=2000, verbose=F)
DefaultAssay(immune.combined)<-'RNA'
immune.combined = FindVariableFeatures(immune.combined, selection.method="vst", nfeatures=2000, verbose=F)
#mask horizontal genes
x=heca.full
masked.genes<-c(
c(grep("^RPL", rownames(x), value = T),grep("^RPS", rownames(x), value = T)),
grep("^MT-", rownames(x), value = T),
c(grep("^IFI", rownames(x), value = T),grep("^ISG", rownames(x), value = T)),
grep("^SMC[0-9]*", rownames(x), value = T),
grep("^MCM[0-9]*", rownames(x), value = T),
c(grep("^TUBA", rownames(x), value = T),
grep("^TUBB", rownames(x), value = T),
grep("^TUBD[0-9]", rownames(x), value = T),
grep("^TUBE[0-9]", rownames(x), value = T),
grep("^TUBG[0-9]", rownames(x), value = T)
),
c(Seurat::cc.genes.updated.2019$s.genes,
Seurat::cc.genes.updated.2019$g2m.genes
),
c("H1-0","H1-1","H1-10","H1-12P","H1-2","H1-3","H1-4","H1-5","H1-6","H1-7","H1-8",
"H1-9P","H2AB1","H2AB2","H2AB3","H2AC1","H2AC10P","H2AC11","H2AC12","H2AC13","H2AC14",
"H2AC15","H2AC16","H2AC17","H2AC18","H2AC19","H2AC20","H2AC21","H2AC2P","H2AC3P","H2AC4",
"H2AC5P","H2AC6","H2AC7","H2AC8","H2AC9P","H2AJ","H2AL1MP","H2AL1Q","H2AL3",
"H2AP","H2AQ1P","H2AW","H2AX","H2AZ1","H2AZ2","MACROH2A1","MACROH2A2","H2BC1","H2BC10",
"H2BC11","H2BC12","H2BC13","H2BC14","H2BC15","H2BC16P","H2BC17","H2BC18",
"H2BC19P","H2BC20P","H2BC21","H2BC2P","H2BC3","H2BC4","H2BC5","H2BC6","H2BC7","H2BC8",
"H2BC9","H2BK1","H2BL1P","H2BN1","H2BC12L","H2BU1","H2BU2P","H2BW1","H2BW2",
"H2BW3P","H2BW4P","H3-7","H3-3A","H3-3B","H3-4","H3-5","H3C1","H3C10","H3C11","H3C12",
"H3C13","H3C14","H3C15","H3C2","H3C3","H3C4","H3C5P","H3C6","H3C7","H3C8",
"H3C9P","H3Y1","H3Y2","CENPA","H4-16","H4C1","H4C10P","H4C11","H4C12","H4C13","H4C14",
"H4C15","H4C2","H4C3","H4C4","H4C5","H4C6","H4C7","H4C8","H4C9"),
grep("^HIST", rownames(x), value=T)
)
goi1 = setdiff( VariableFeatures(heca.full), masked.genes)
# mask horizontal genes
x=immune.combined
masked.genes<-c(
c(grep("^RPL", rownames(x), value = T),grep("^RPS", rownames(x), value = T)),
grep("^MT-", rownames(x), value = T),
c(grep("^IFI", rownames(x), value = T),grep("^ISG", rownames(x), value = T)),
grep("^SMC[0-9]*", rownames(x), value = T),
grep("^MCM[0-9]*", rownames(x), value = T),
c(grep("^TUBA", rownames(x), value = T),
grep("^TUBB", rownames(x), value = T),
grep("^TUBD[0-9]", rownames(x), value = T),
grep("^TUBE[0-9]", rownames(x), value = T),
grep("^TUBG[0-9]", rownames(x), value = T)
),
c(Seurat::cc.genes.updated.2019$s.genes,
Seurat::cc.genes.updated.2019$g2m.genes
),
c("H1-0","H1-1","H1-10","H1-12P","H1-2","H1-3","H1-4","H1-5","H1-6","H1-7","H1-8",
"H1-9P","H2AB1","H2AB2","H2AB3","H2AC1","H2AC10P","H2AC11","H2AC12","H2AC13","H2AC14",
"H2AC15","H2AC16","H2AC17","H2AC18","H2AC19","H2AC20","H2AC21","H2AC2P","H2AC3P","H2AC4",
"H2AC5P","H2AC6","H2AC7","H2AC8","H2AC9P","H2AJ","H2AL1MP","H2AL1Q","H2AL3",
"H2AP","H2AQ1P","H2AW","H2AX","H2AZ1","H2AZ2","MACROH2A1","MACROH2A2","H2BC1","H2BC10",
"H2BC11","H2BC12","H2BC13","H2BC14","H2BC15","H2BC16P","H2BC17","H2BC18",
"H2BC19P","H2BC20P","H2BC21","H2BC2P","H2BC3","H2BC4","H2BC5","H2BC6","H2BC7","H2BC8",
"H2BC9","H2BK1","H2BL1P","H2BN1","H2BC12L","H2BU1","H2BU2P","H2BW1","H2BW2",
"H2BW3P","H2BW4P","H3-7","H3-3A","H3-3B","H3-4","H3-5","H3C1","H3C10","H3C11","H3C12",
"H3C13","H3C14","H3C15","H3C2","H3C3","H3C4","H3C5P","H3C6","H3C7","H3C8",
"H3C9P","H3Y1","H3Y2","CENPA","H4-16","H4C1","H4C10P","H4C11","H4C12","H4C13","H4C14",
"H4C15","H4C2","H4C3","H4C4","H4C5","H4C6","H4C7","H4C8","H4C9"),
grep("^HIST", rownames(x), value=T)
)
goi2 = setdiff( VariableFeatures(immune.combined), masked.genes)
goi= unique(c(goi1,goi2))
length(goi)
plan("sequential")
options(future.globals.maxSize = 1000 * 1024^2*4)
plan("multiprocess", workers = 14)
heca.full <- ScaleData(heca.full, features = goi)
immune.combined <- ScaleData(immune.combined, features = goi)
plan("sequential")
Warning message:
“Strategy 'multiprocess' is deprecated in future (>= 1.20.0). Instead, explicitly specify either 'multisession' or 'multicore'. In the current R session, 'multiprocess' equals 'multicore'.”
Centering and scaling data matrix
Centering and scaling data matrix
plan("sequential")
options(future.globals.maxSize = 1000 * 1024^2*4)
plan("multiprocess", workers = 14)
transfer.anchors <- FindTransferAnchors(reference = heca.full, query = immune.combined,
dims = 1:30, features=goi)
plan("sequential")
Warning message:
“844 features of the features specified were not present in both the reference query assays.
Continuing with remaining 2486 features.”
Performing PCA on the provided reference using 2486 features as input.
Projecting cell embeddings
Finding neighborhoods
Finding anchors
Found 16065 anchors
Filtering anchors
Retained 6000 anchors
predictions <- TransferData(anchorset = transfer.anchors, refdata = heca.full$ann,
dims = 1:30)
immune.combined <- AddMetaData(immune.combined, metadata = predictions)
Finding integration vectors
Finding integration vector weights
Predicting cell labels
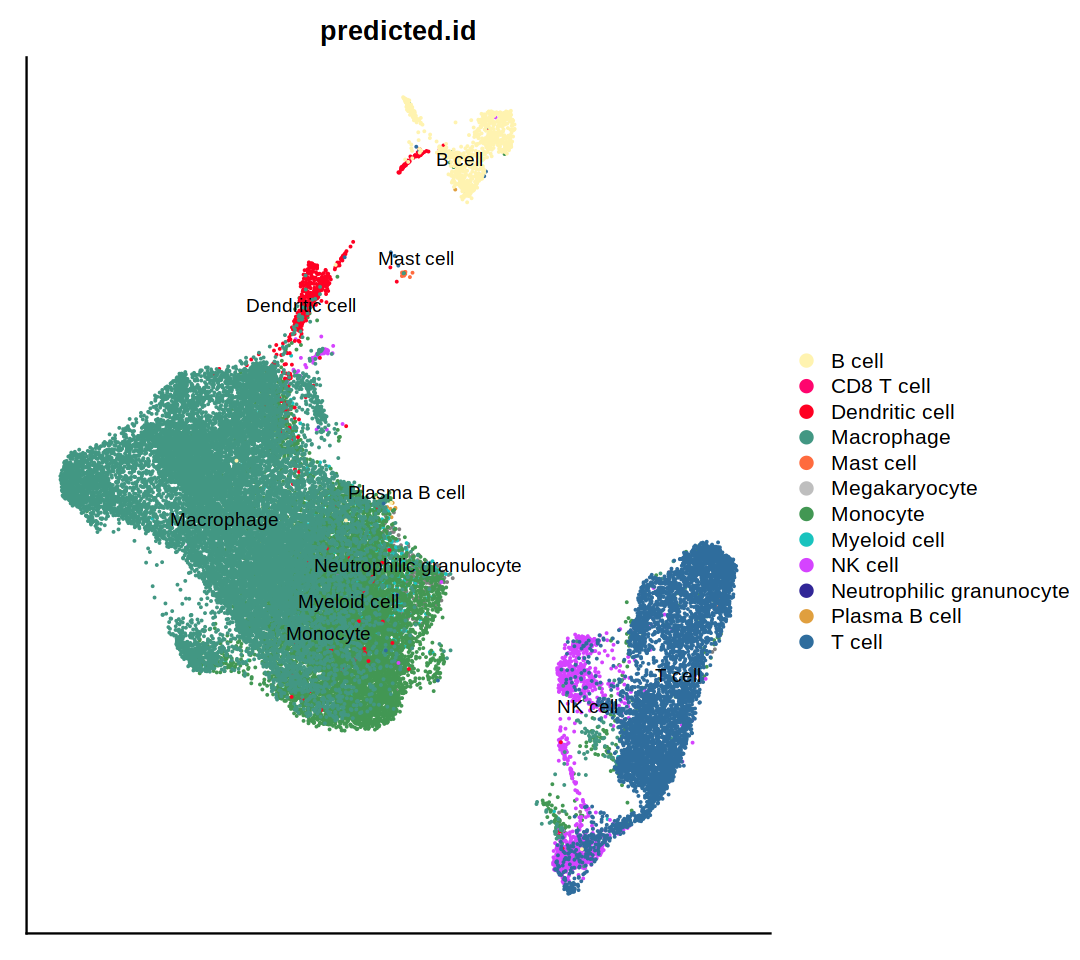
o(10,8)
DimPlot(immune.combined, group.by='predicted.id', label=T, repel=T, cols=heca.color.table, order='Neutrophilic granunocyte')
DimPlot(immune.combined, group.by='persample_ann', label=T, repel=T, cols=covid.color.table)
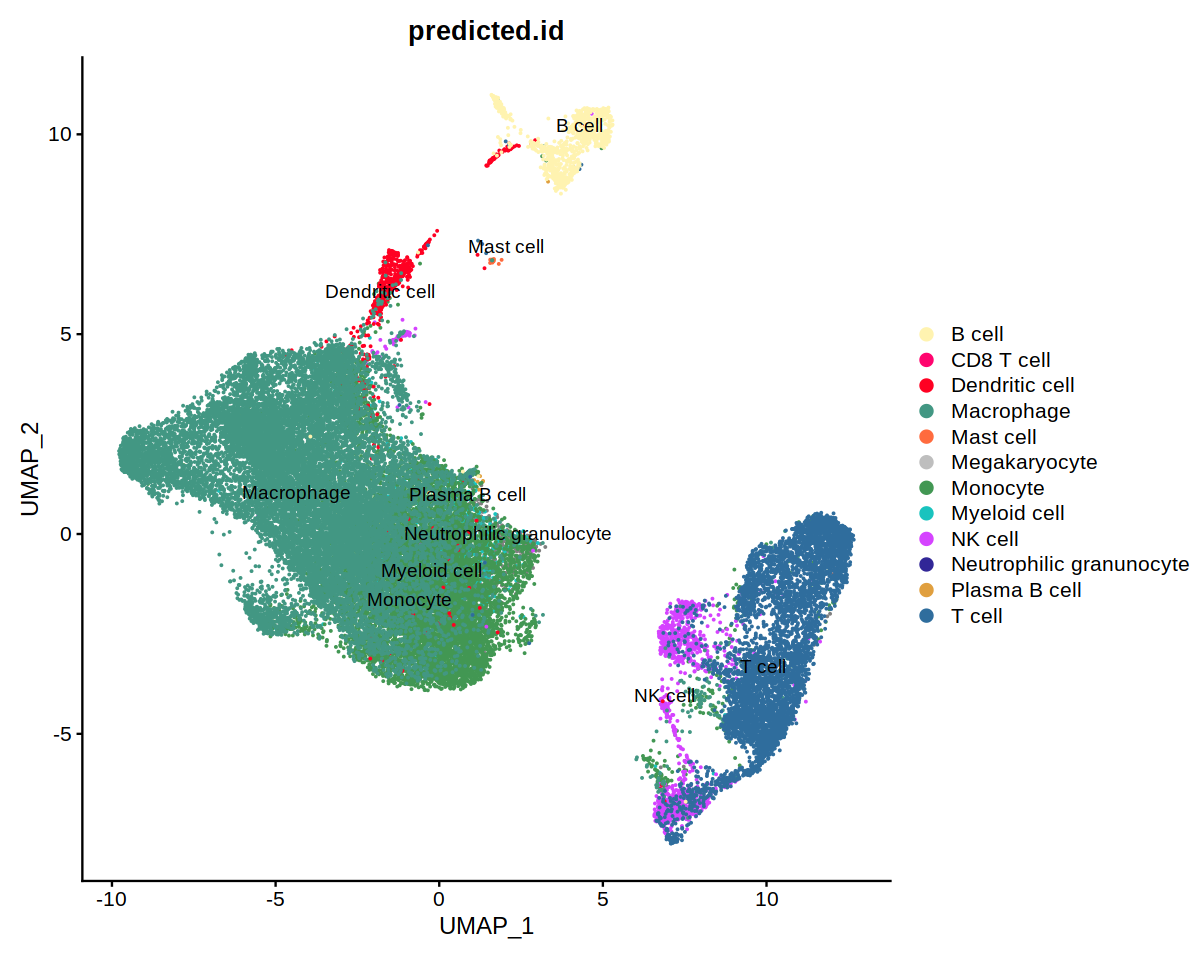
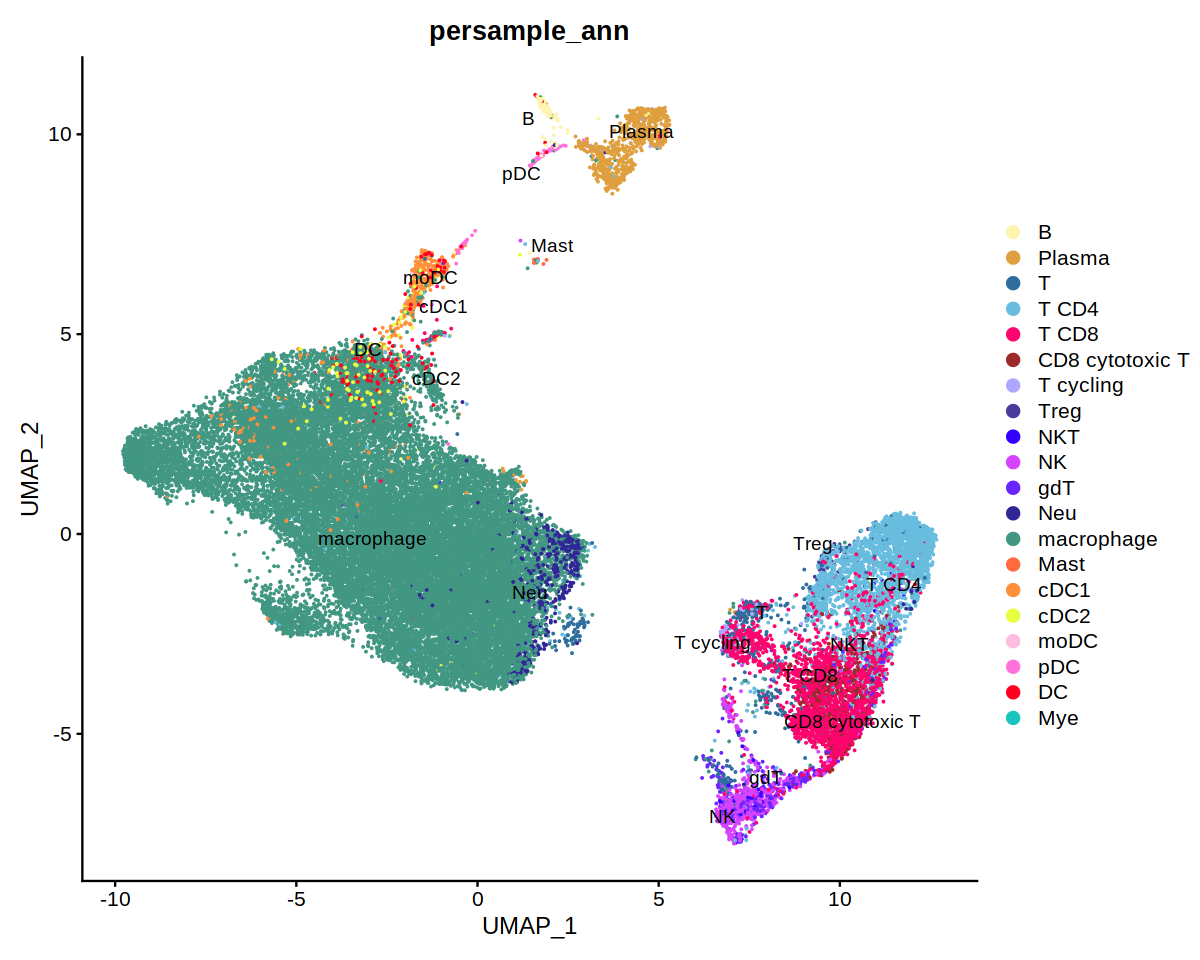
o(9,8)
DimPlot(immune.combined, group.by='predicted.id', label=T, repel=T, cols=heca.color.table, order='Neutrophilic granunocyte')&
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank())
cairo_pdf(filename = 'covid19.umap.predicted.pdf', width = 9,height = 8)
DimPlot(immune.combined, group.by='predicted.id', label=T, repel=T, cols=heca.color.table, order='Neutrophilic granunocyte')&
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank())
dev.off()
matched <- c(
paste('B' , 'B cell', sep="_"),
paste('Plasma' , 'Plasma B cell', sep="_"),
paste('Plasma' , 'B cell', sep="_"),
paste('T' , 'CD8 T cell', sep="_"),
paste('T' , 'T cell', sep="_"),
paste('T CD4' , 'T cell', sep="_"),
paste('T CD8' , 'T cell', sep="_"),
paste('T CD8' , 'CD8 T cell', sep="_"),
paste('CD8 cytotoxic T' , 'T cell', sep="_"),
paste('CD8 cytotoxic T' , 'CD8 T cell', sep="_"),
paste('T cycling' , 'T cell', sep="_"),
paste('T cycling' , 'CD8 T cell', sep="_"),
paste('Treg' , 'T cell', sep="_"),
paste('gdT' , 'T cell', sep="_"),
paste('NKT' , 'T cell', sep="_"),
paste('NK' , 'NK cell', sep="_"),
paste('Neu' , 'Neutrophilic granulocyte', sep="_"),
paste('macrophage' , 'Macrophage', sep="_"),
paste('macrophage' , 'Monocyte', sep="_"),
paste('macrophage' , 'Myeloid cell', sep="_"),
paste('Mast' , 'Mast cell', sep="_"),
paste('cDC1' , 'Dendritic cell', sep="_"),
paste('cDC1' , 'Myeloid cell', sep="_"),
paste('cDC2' , 'Dendritic cell', sep="_"),
paste('cDC2' , 'Myeloid cell', sep="_"),
paste('moDC' , 'Dendritic cell', sep="_"),
paste('pDC' , 'Dendritic cell', sep="_"),
paste('DC' , 'Dendritic cell', sep="_"),
paste('Mye' , 'Macrophage', sep="_"),
paste('Mye' , 'Mast cell', sep="_"),
paste('Mye' , 'Megakaryocyte', sep="_"),
paste('Mye' , 'Dendritic cell', sep="_"),
paste('Mye' , 'Monocyte', sep="_"),
paste('Mye' , 'Neutrophilic granulocyte', sep="_"),
paste('Mye' , 'Myeloid cell', sep="_")
)
# calculate accuracy
meta=immune.combined@meta.data
meta = meta[, c('persample_ann','predicted.id')]
meta$joint <- paste(meta$persample_ann, meta$predicted.id, sep='_')
meta$isMatched <- (meta$joint %in% matched) | (meta$persample_ann == meta$predicted.id)
acc = (meta %>% filter(isMatched==T) %>% nrow) / (meta %>% nrow)
print( acc )
[1] 0.9204989
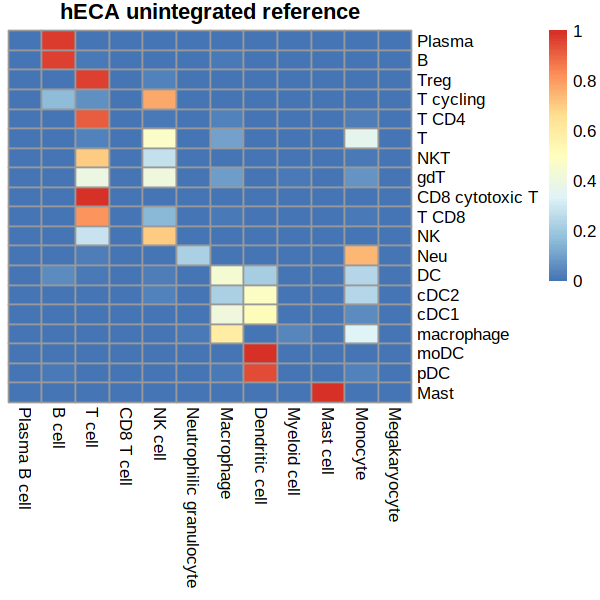
# draw matrix
## create an empty adj matrix
rows = as.character( unique(immune.combined@meta.data$persample_ann) ) # from:covid ground-truth
cols = as.character( unique(heca.full@meta.data$cell_type) ) # to:predicted to
mat <- matrix(0, length(rows), length(cols))
rownames(mat) <- rows
colnames(mat) <- cols
# get an edge list
edge.list = meta %>% count(persample_ann, predicted.id, sort = TRUE)
# fill in the edge list
for(i in 1:NROW(edge.list)) mat[ edge.list[i,1], edge.list[i,2] ] <- edge.list[i,3] # SEE UPDATE
row.order= c('Plasma','B',
'Treg','T cycling','T CD4','T','NKT','gdT','CD8 cytotoxic T',
'T CD8','NK',
'Neu','DC','cDC2','cDC1','macrophage','moDC','pDC','Mast'
)
col.order=c('Plasma B cell','B cell',
'T cell','CD8 T cell','NK cell',
'Neutrophilic granulocyte',
'Macrophage','Dendritic cell','Myeloid cell','Mast cell','Monocyte','Megakaryocyte')
# row-sum scaling
mat = mat/rowSums(mat)
mat = mat[row.order,col.order]
library(pheatmap)
o(5,5)
pheatmap(mat, cluster_rows = F, cluster_cols = F,main='hECA unintegrated reference')
library('mclust')
adjustedRandIndex(meta$persample_ann, meta$predicted.id)
Package 'mclust' version 5.4.9
Type 'citation("mclust")' for citing this R package in publications.