3. Covid19 data QC¶
library(tictoc)
tic()
load(file = 'nCoV.list.rda')
toc()
112.838 sec elapsed
library(Seurat)
library(dplyr)
library(ggpubr)
o<-function(w,h) options(repr.plot.width=w, repr.plot.height=h)
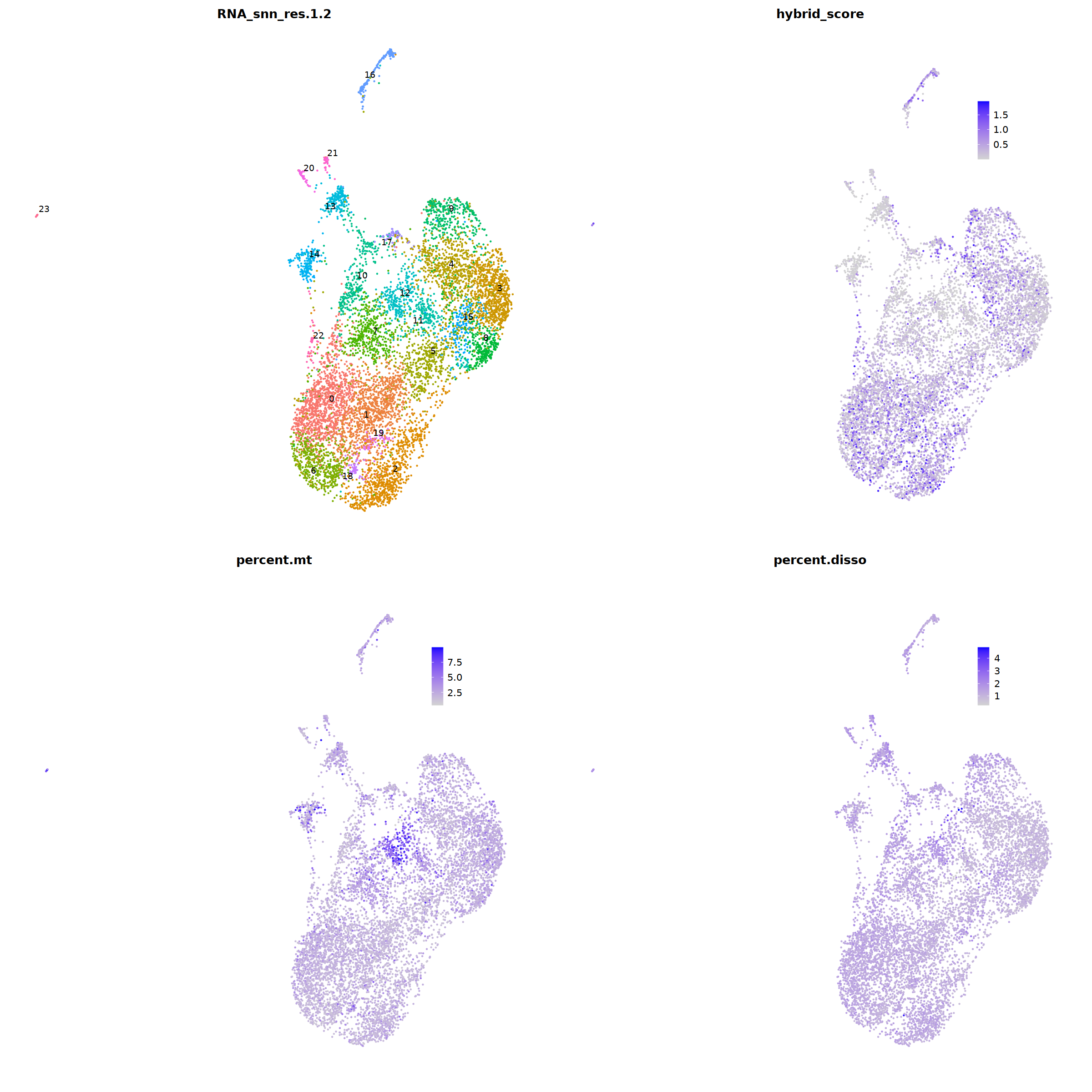
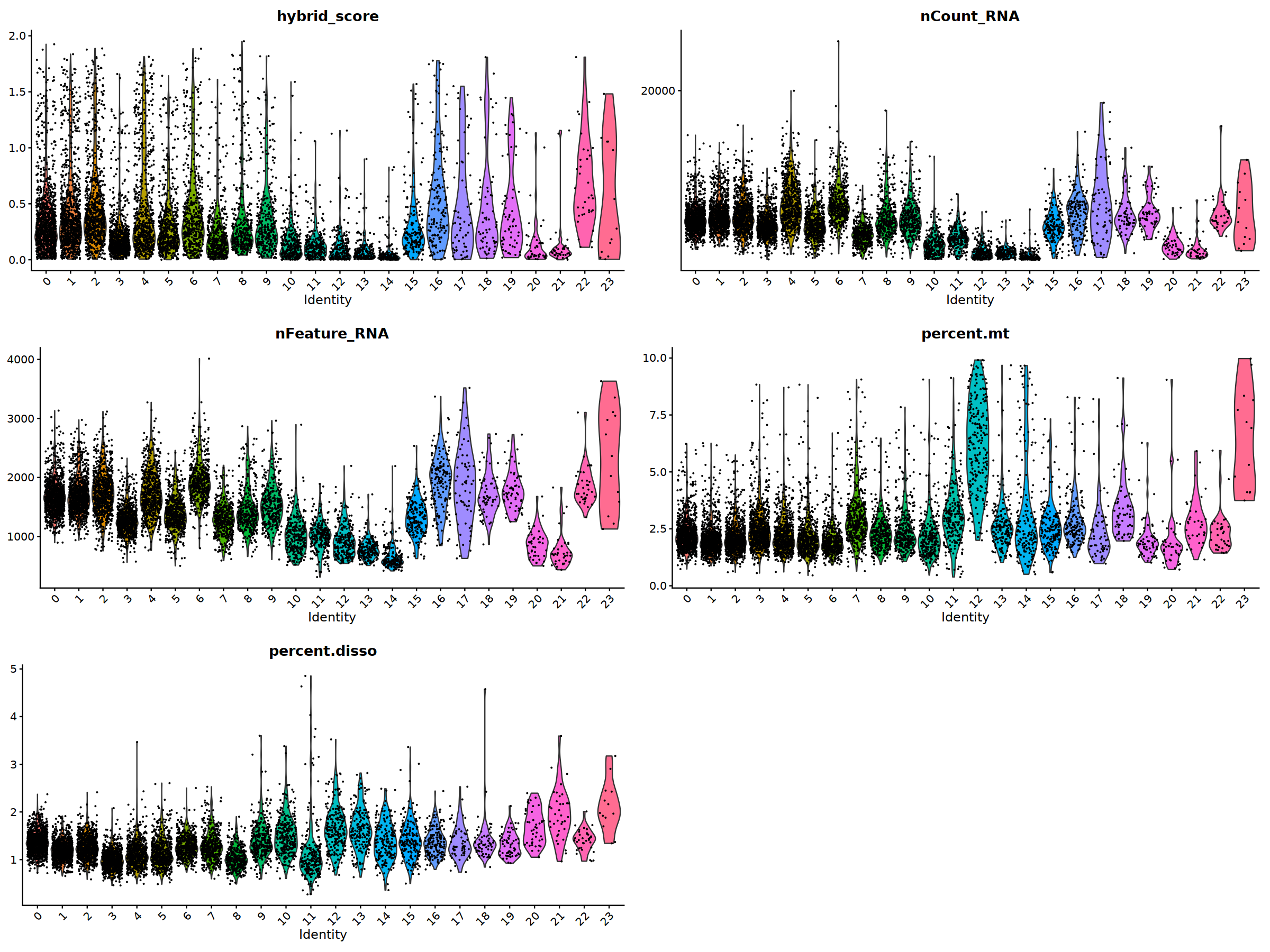
sample 1¶
i=1
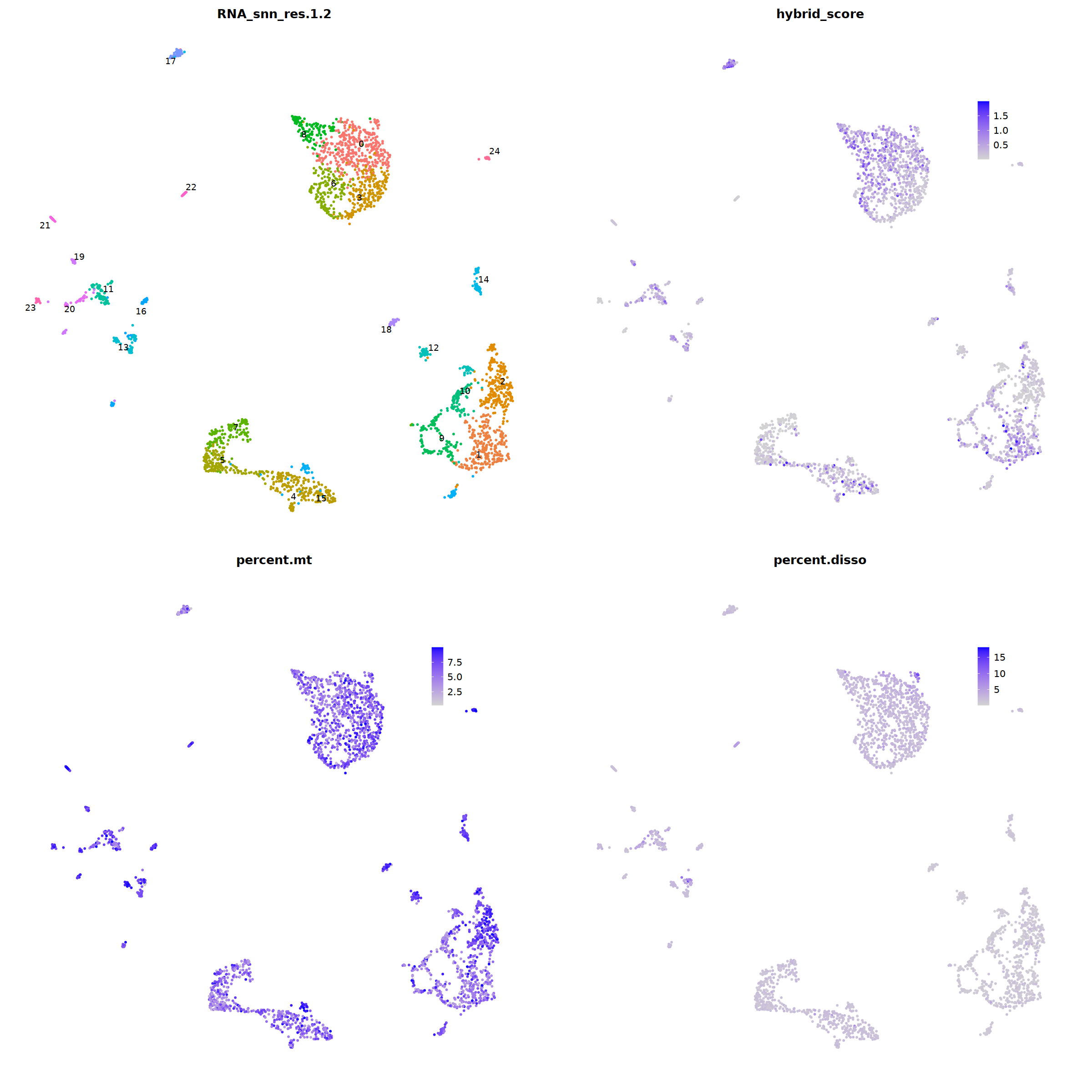
o(20,20)
ggarrange(
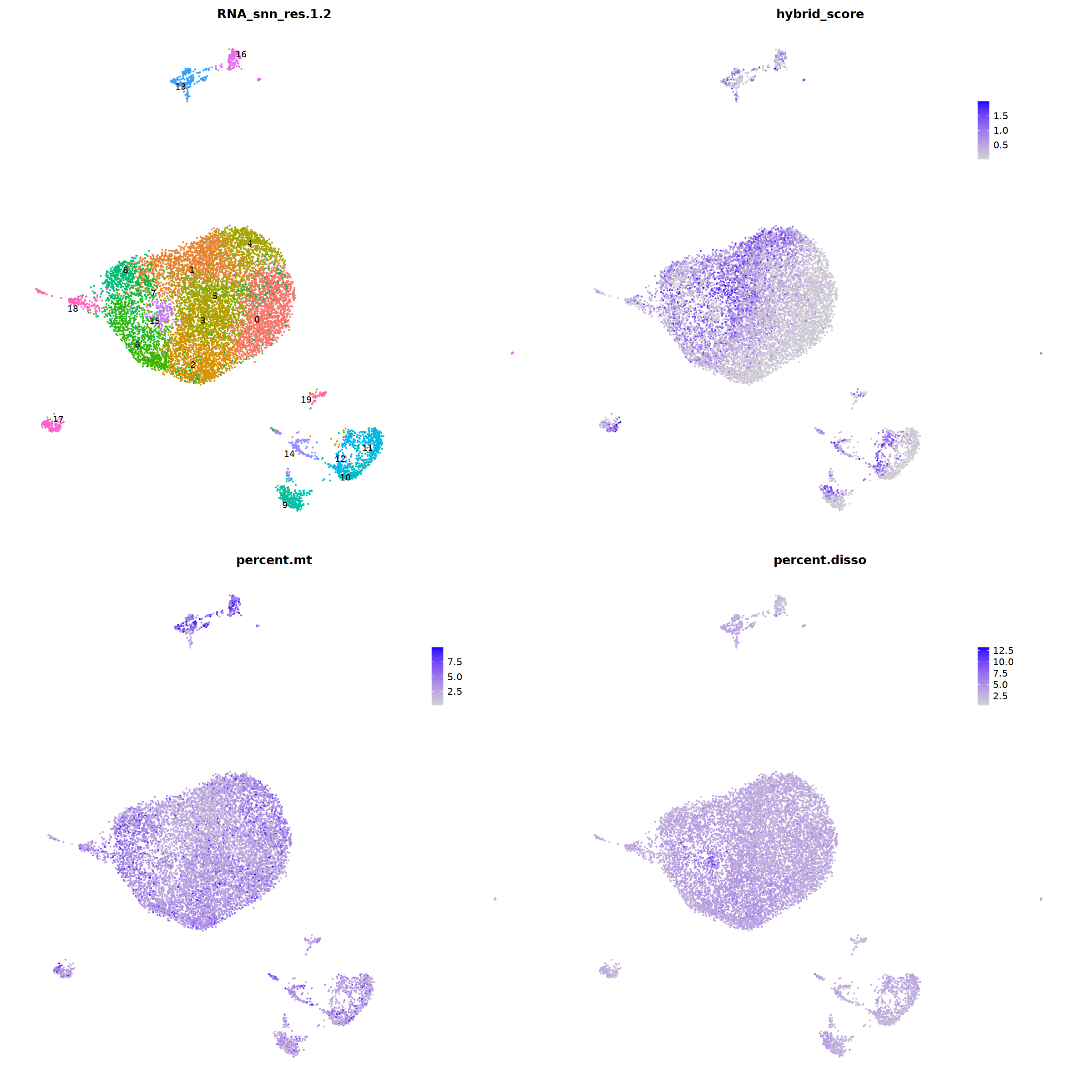
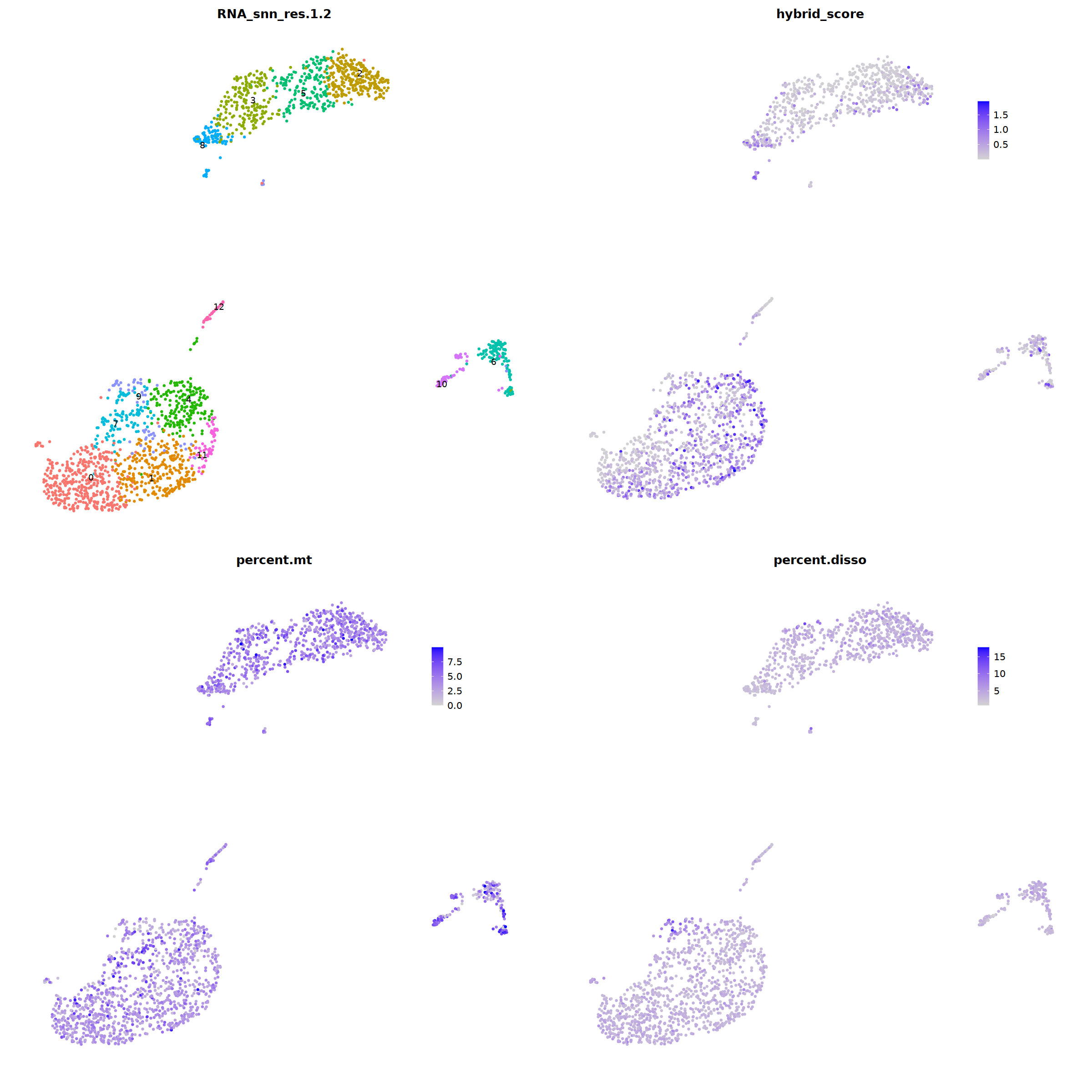
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
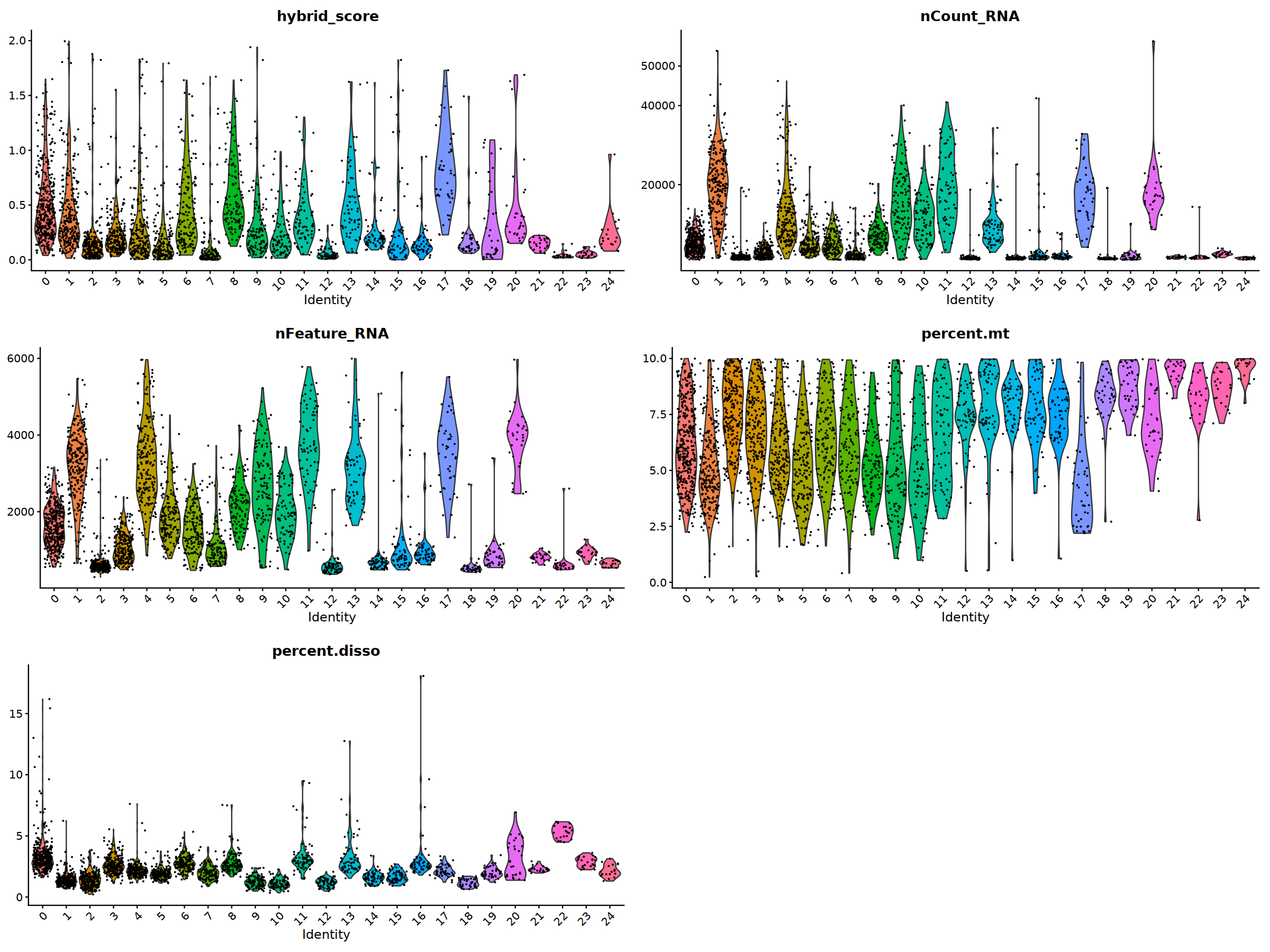
o(20,15)
ggarrange(
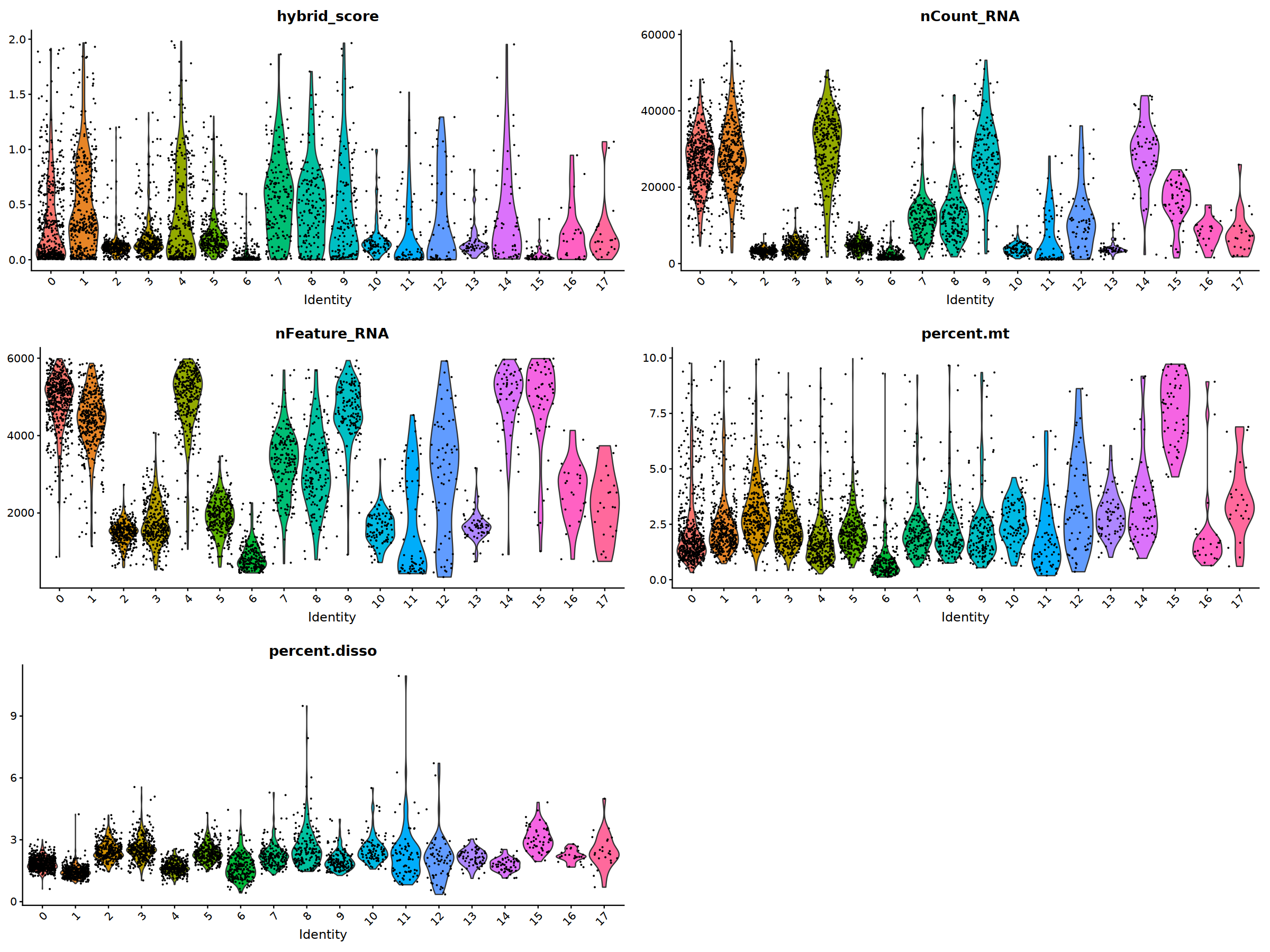
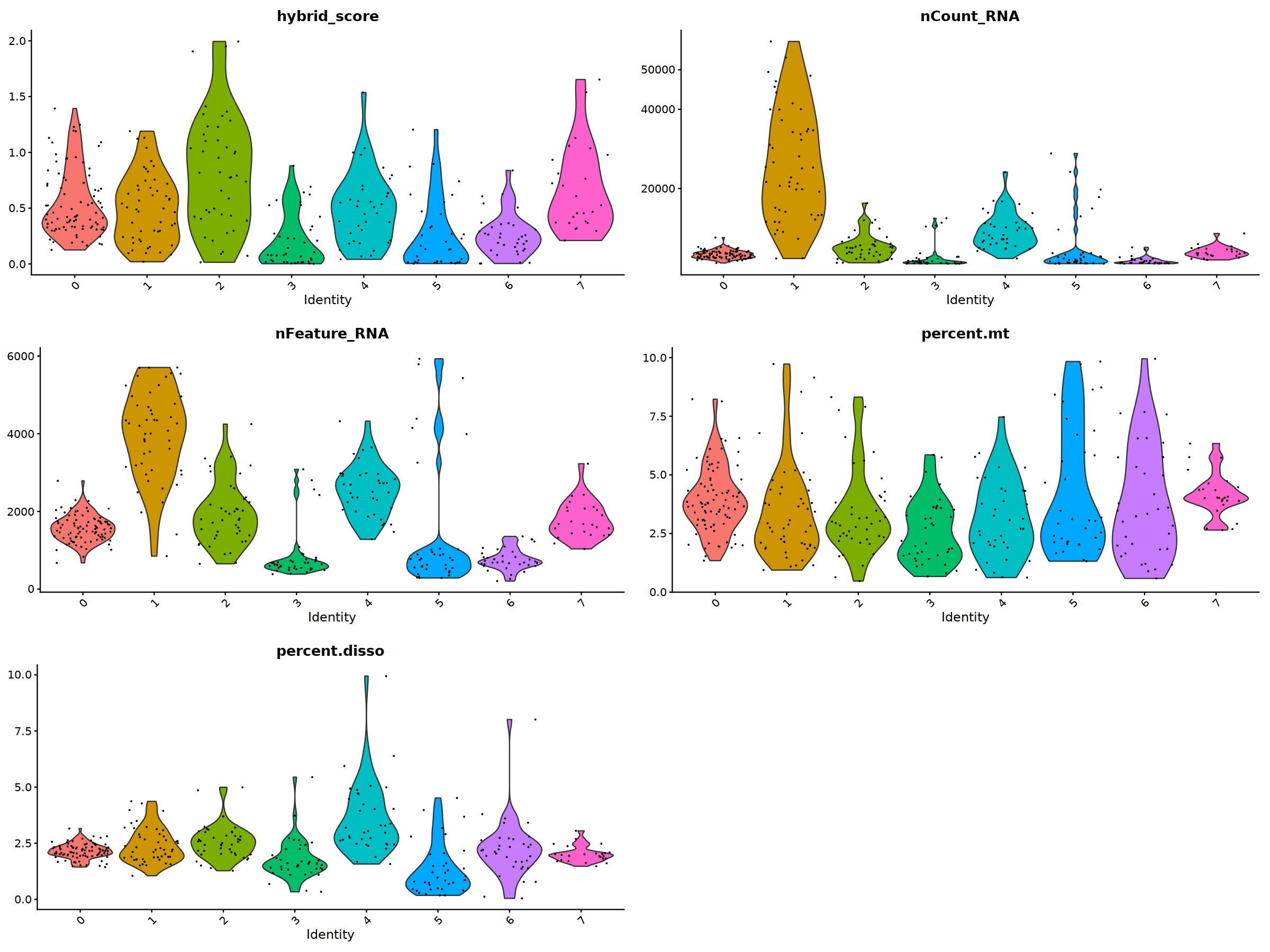
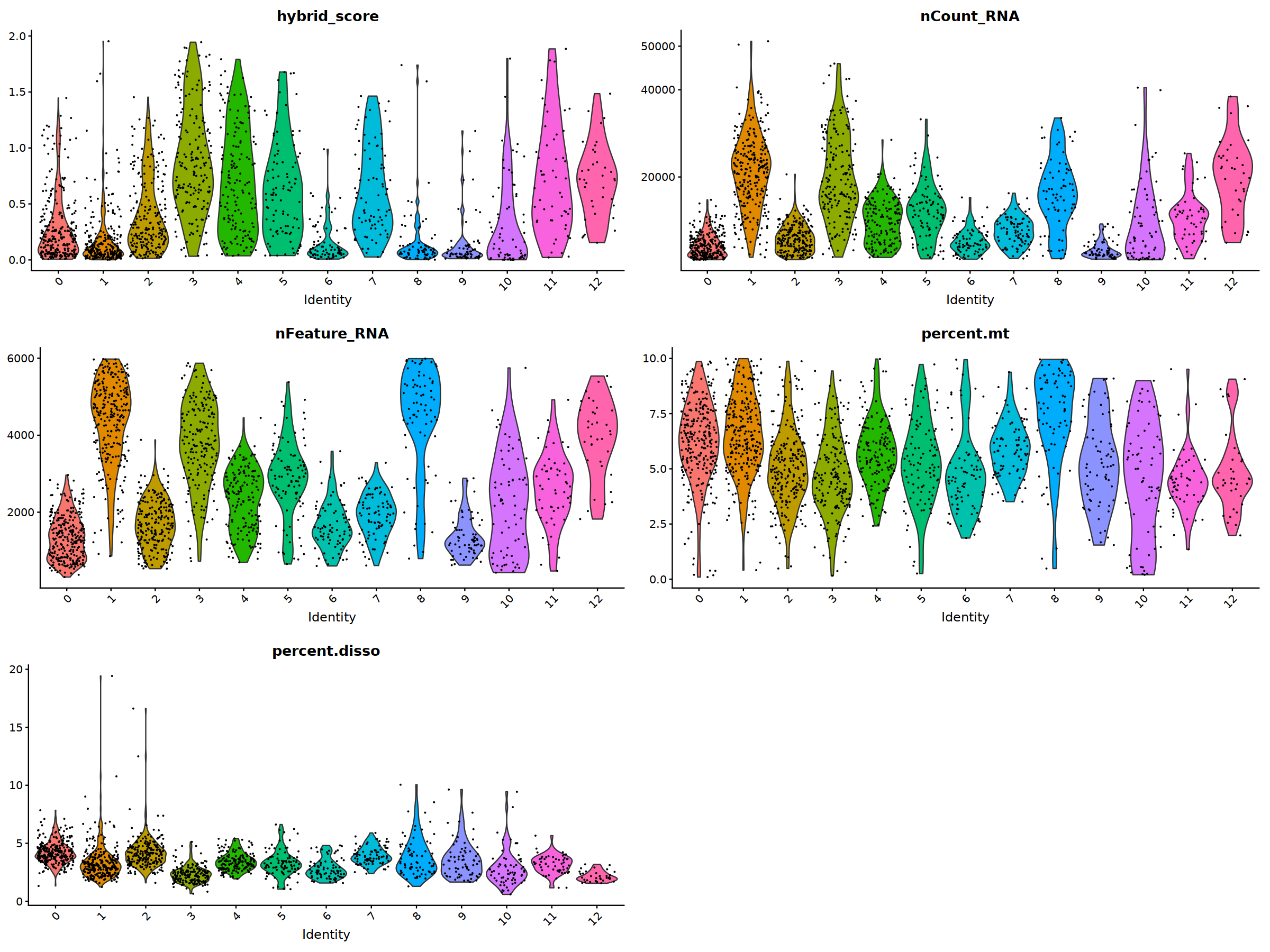
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
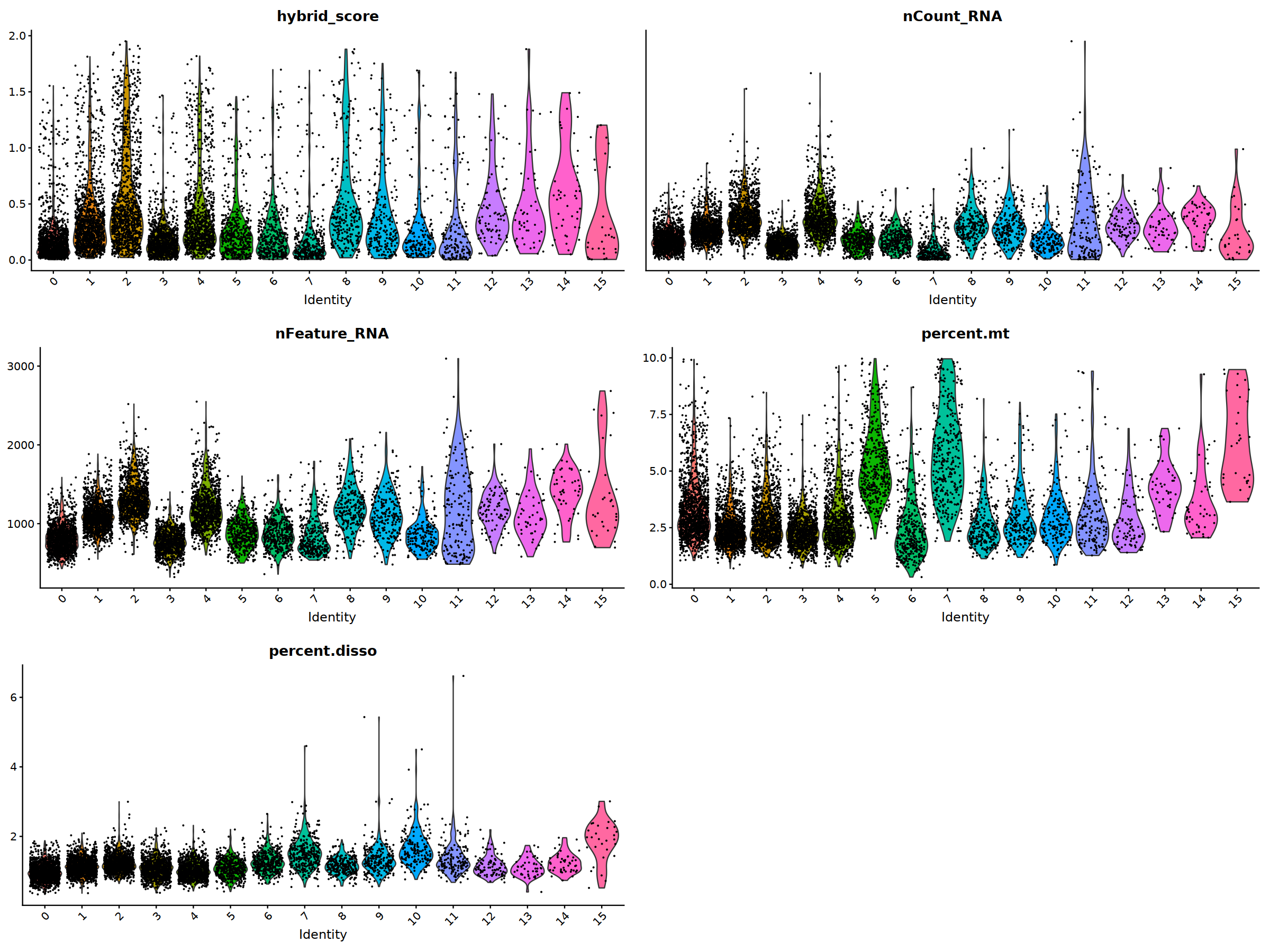
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
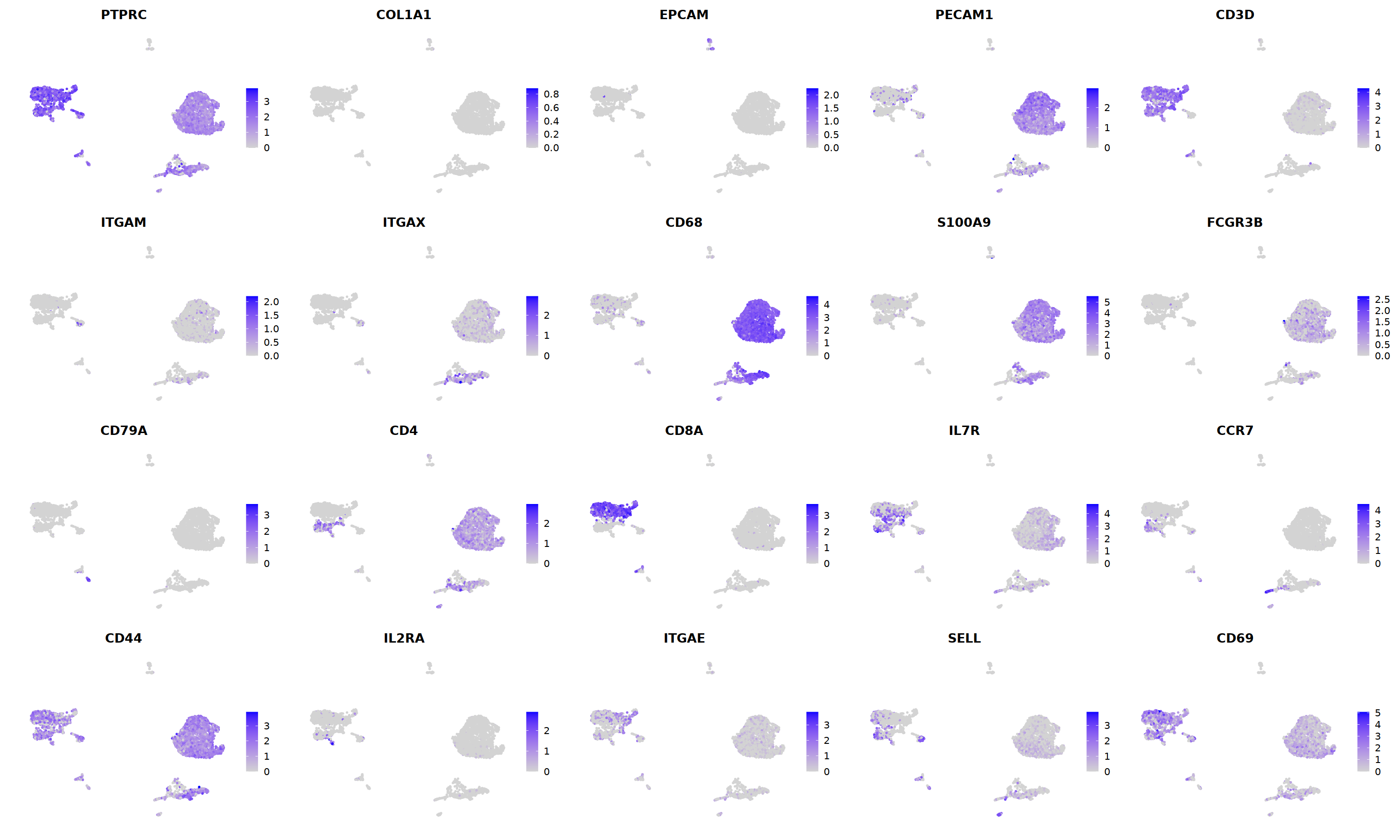
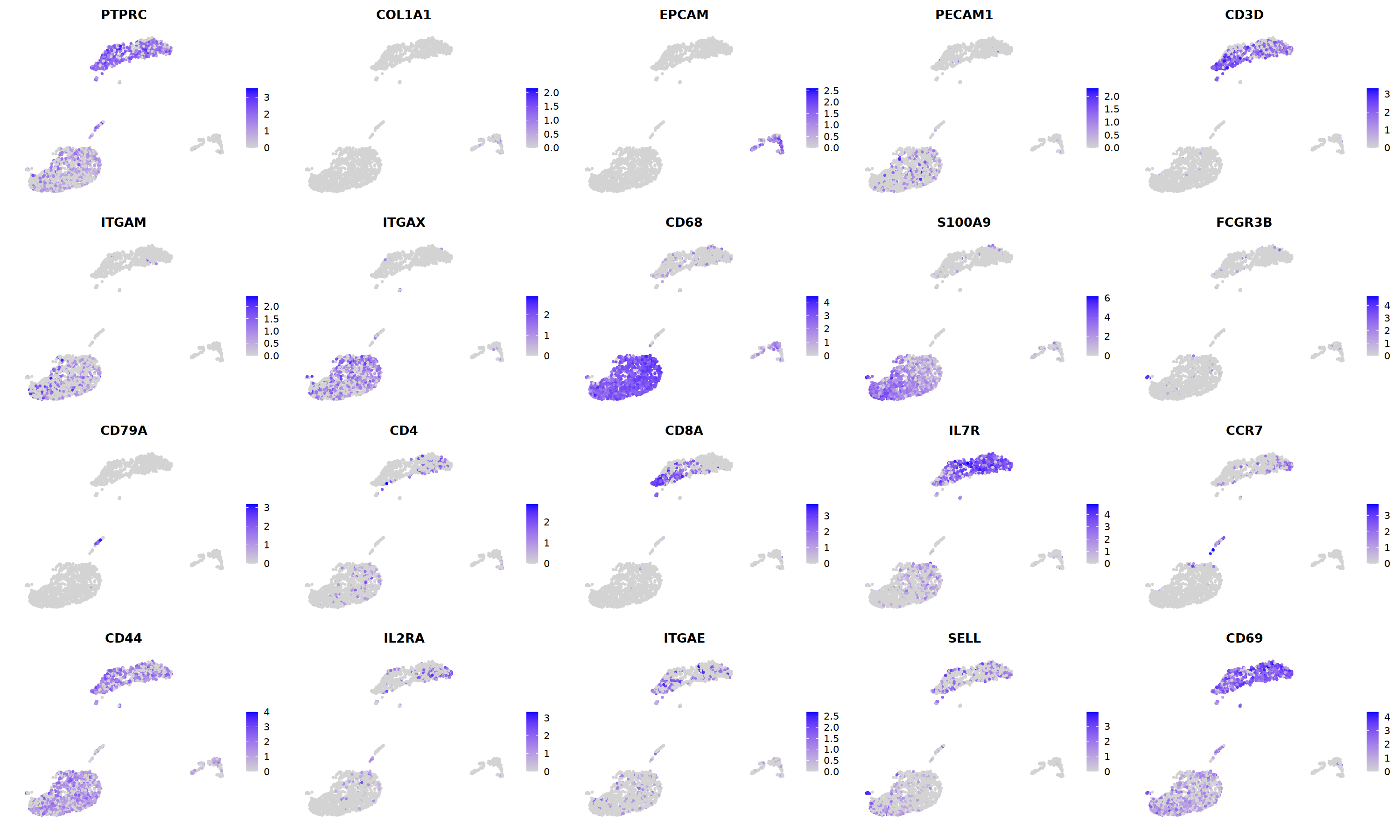
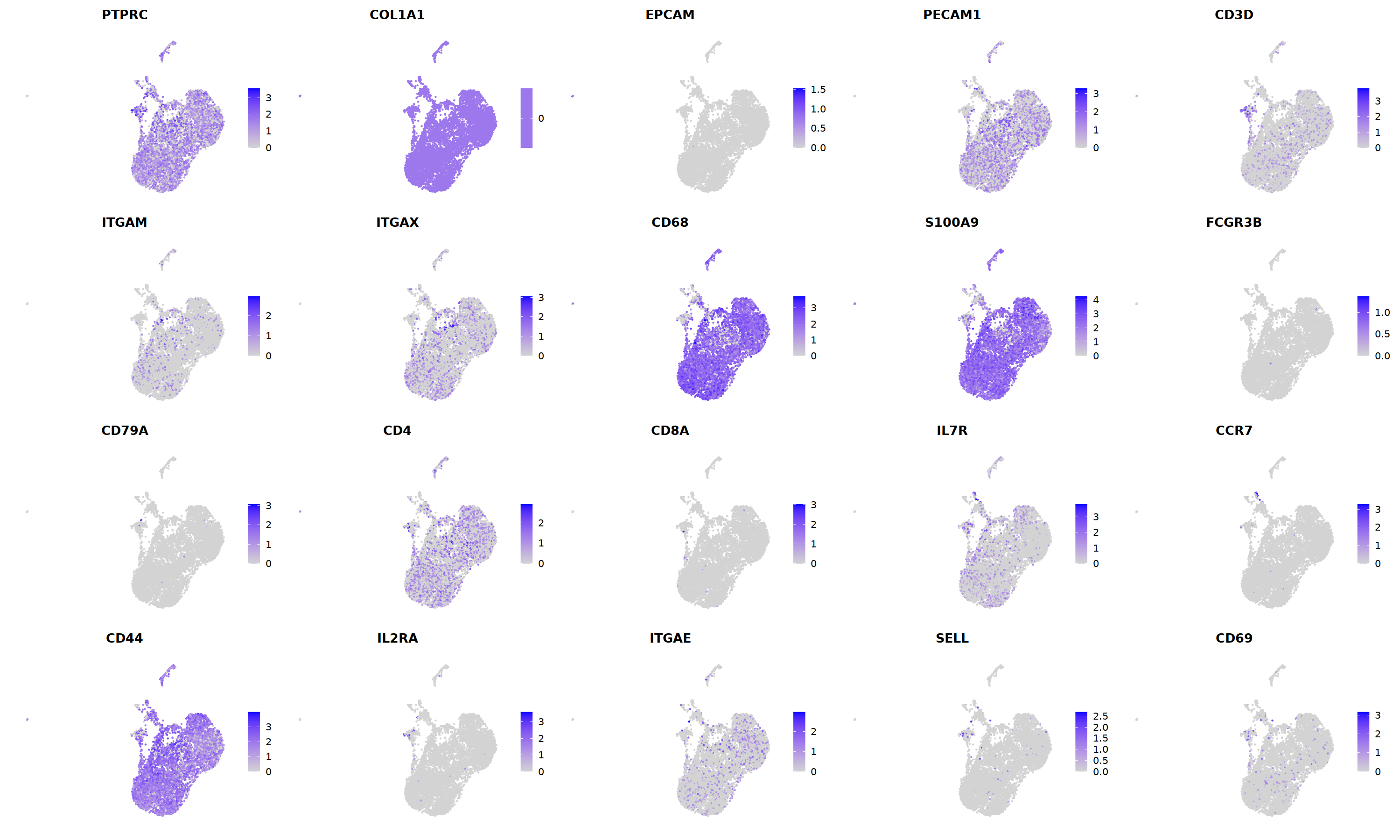
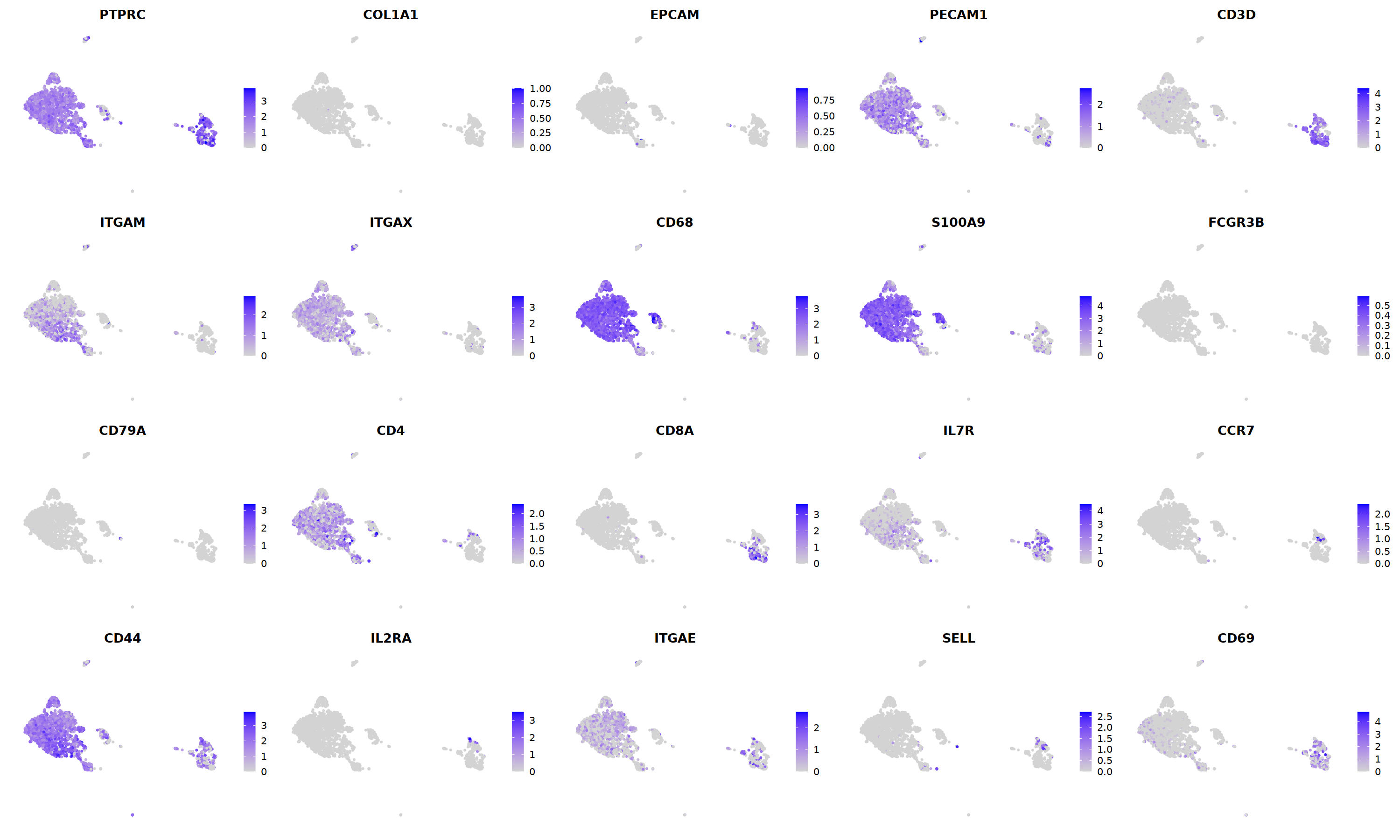
o(5*5, 5*3)
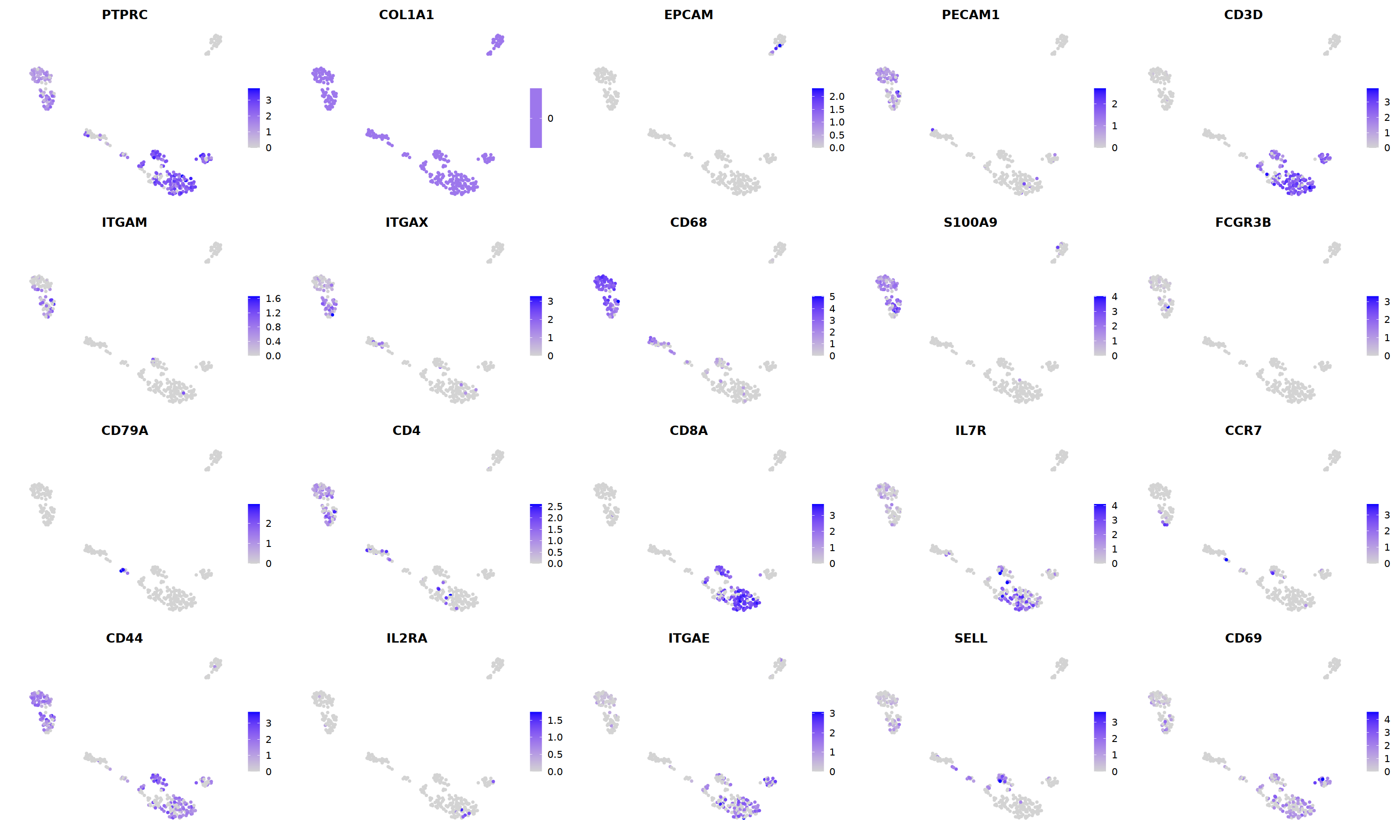
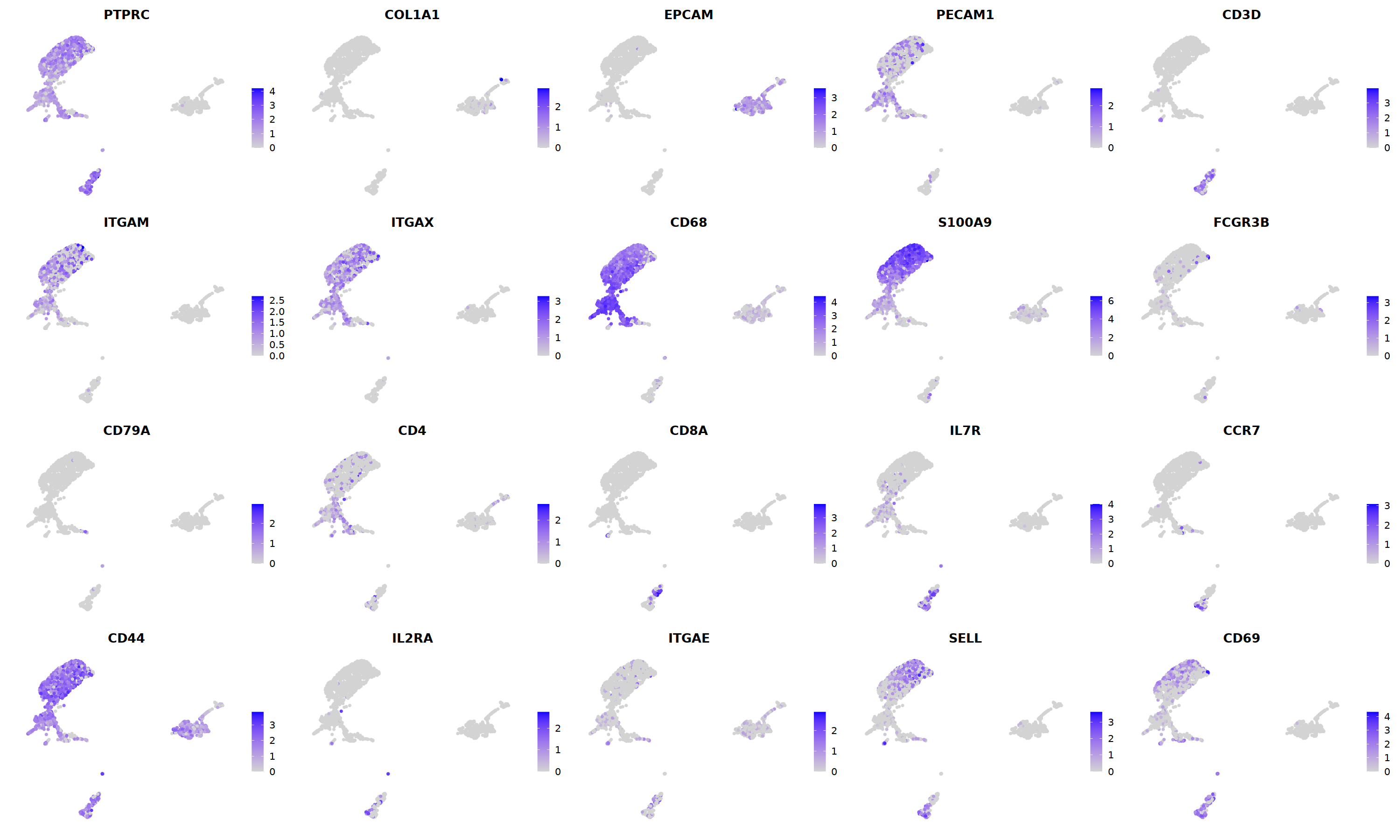
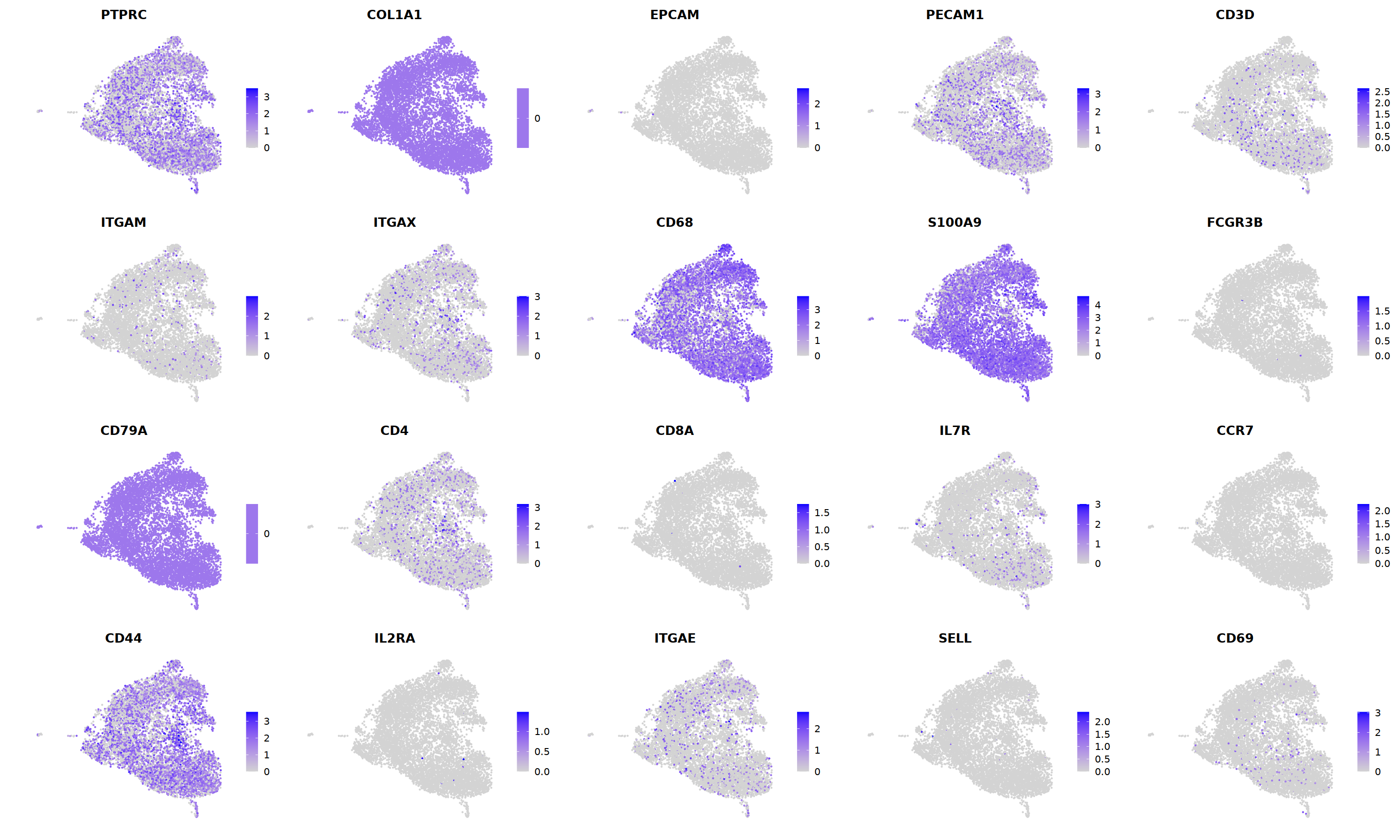
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
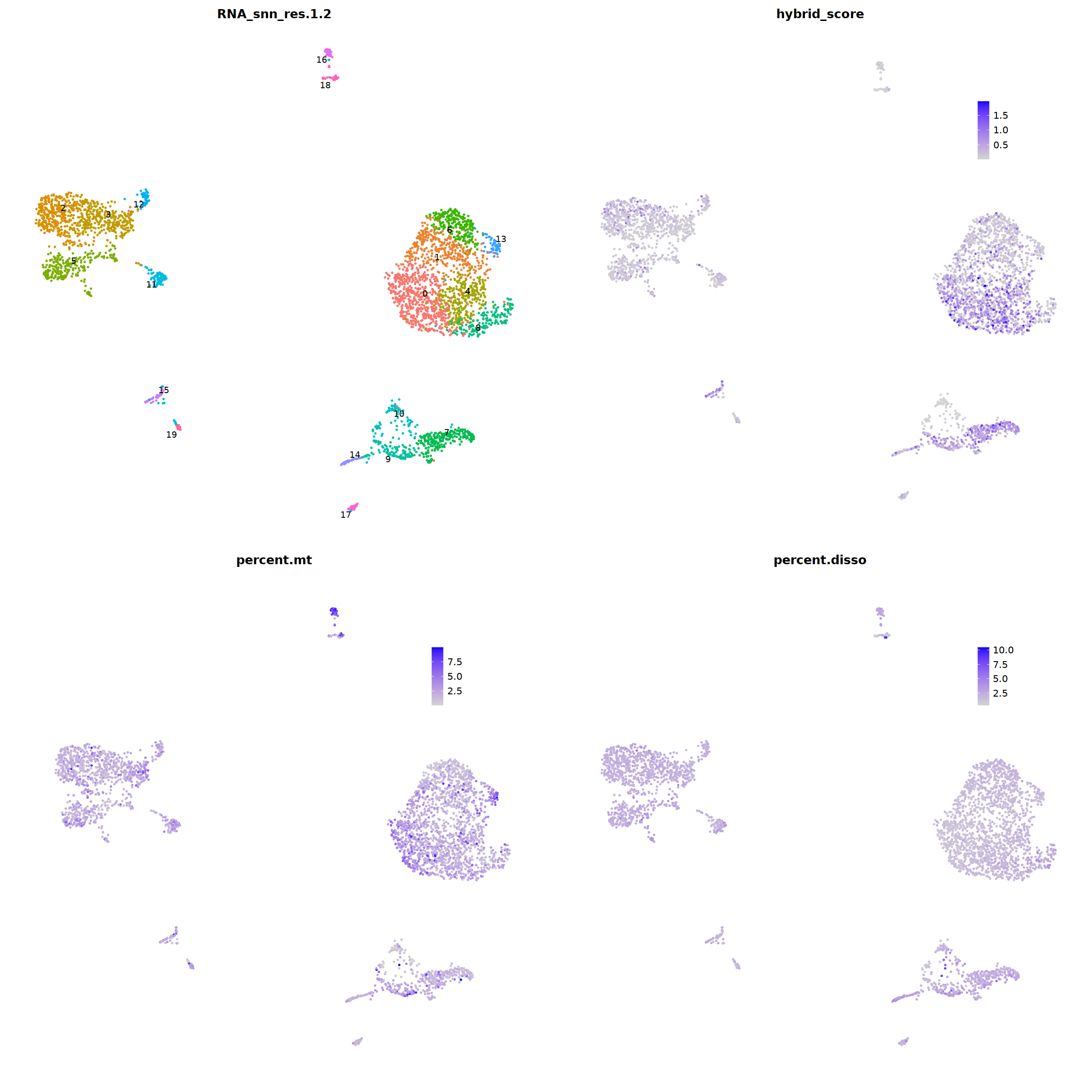
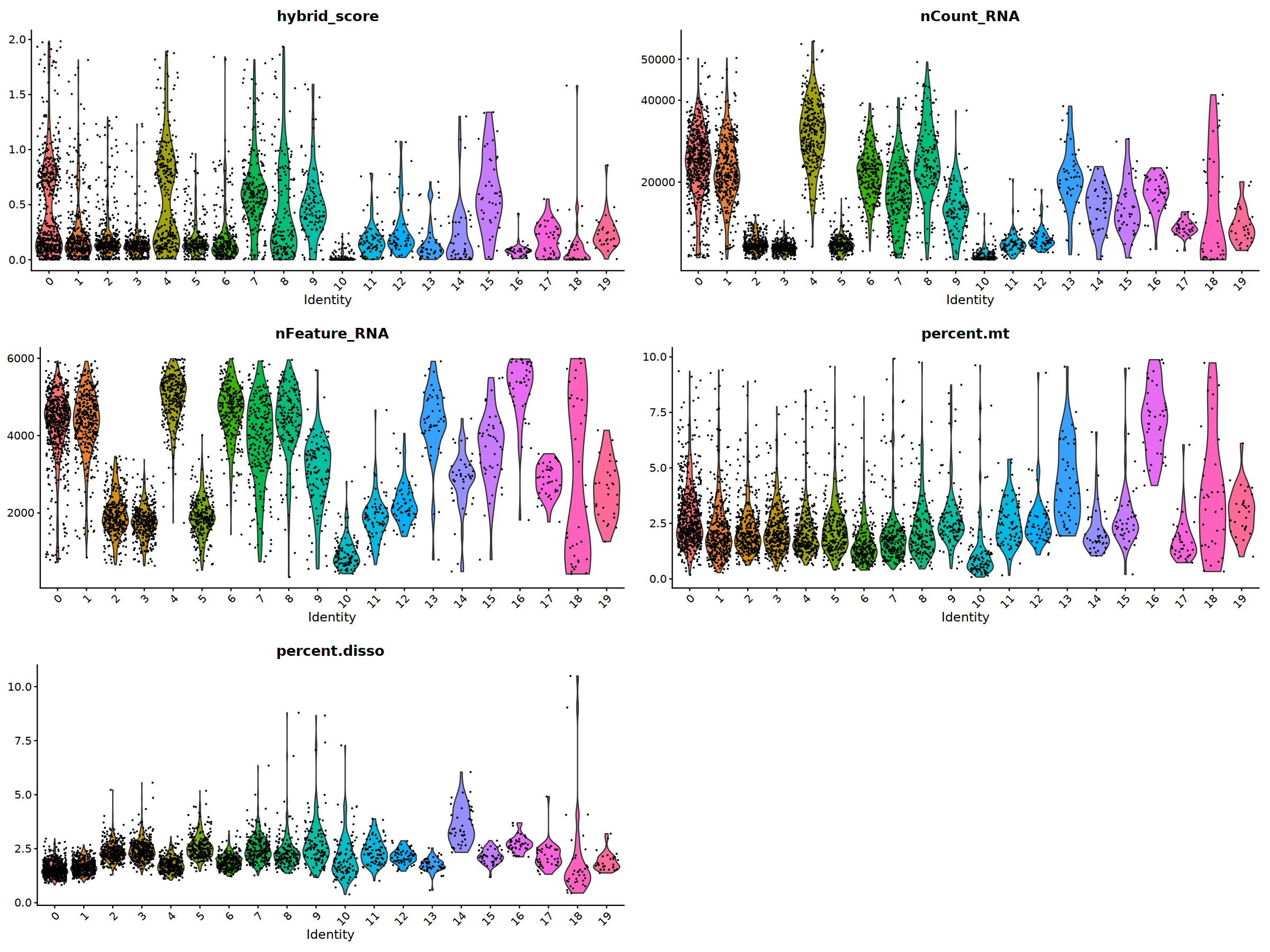
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5)
sample 2¶
i=2
o(20,20)
ggarrange(
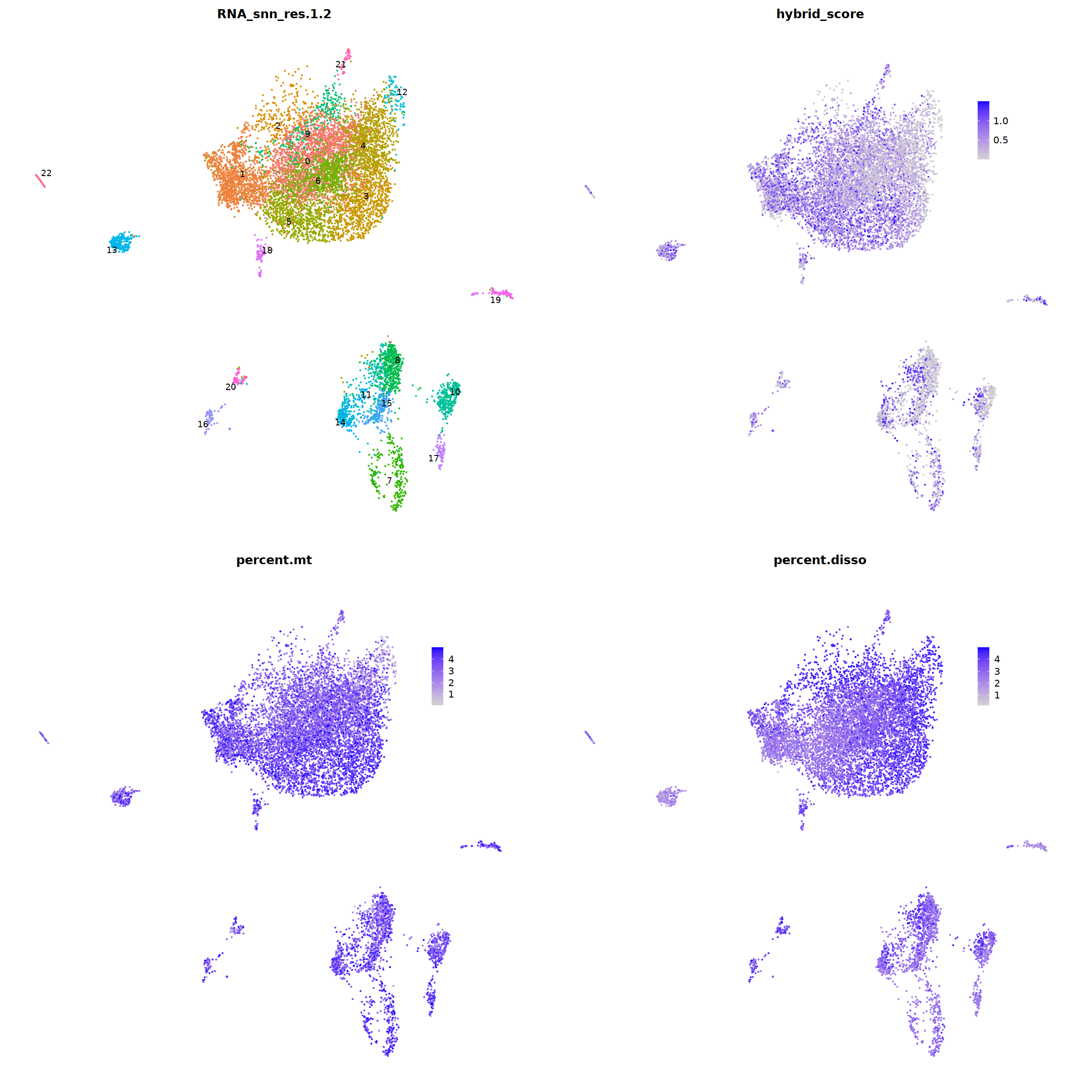
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.mt < 5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5)
sample 3¶
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.mt < 5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5)
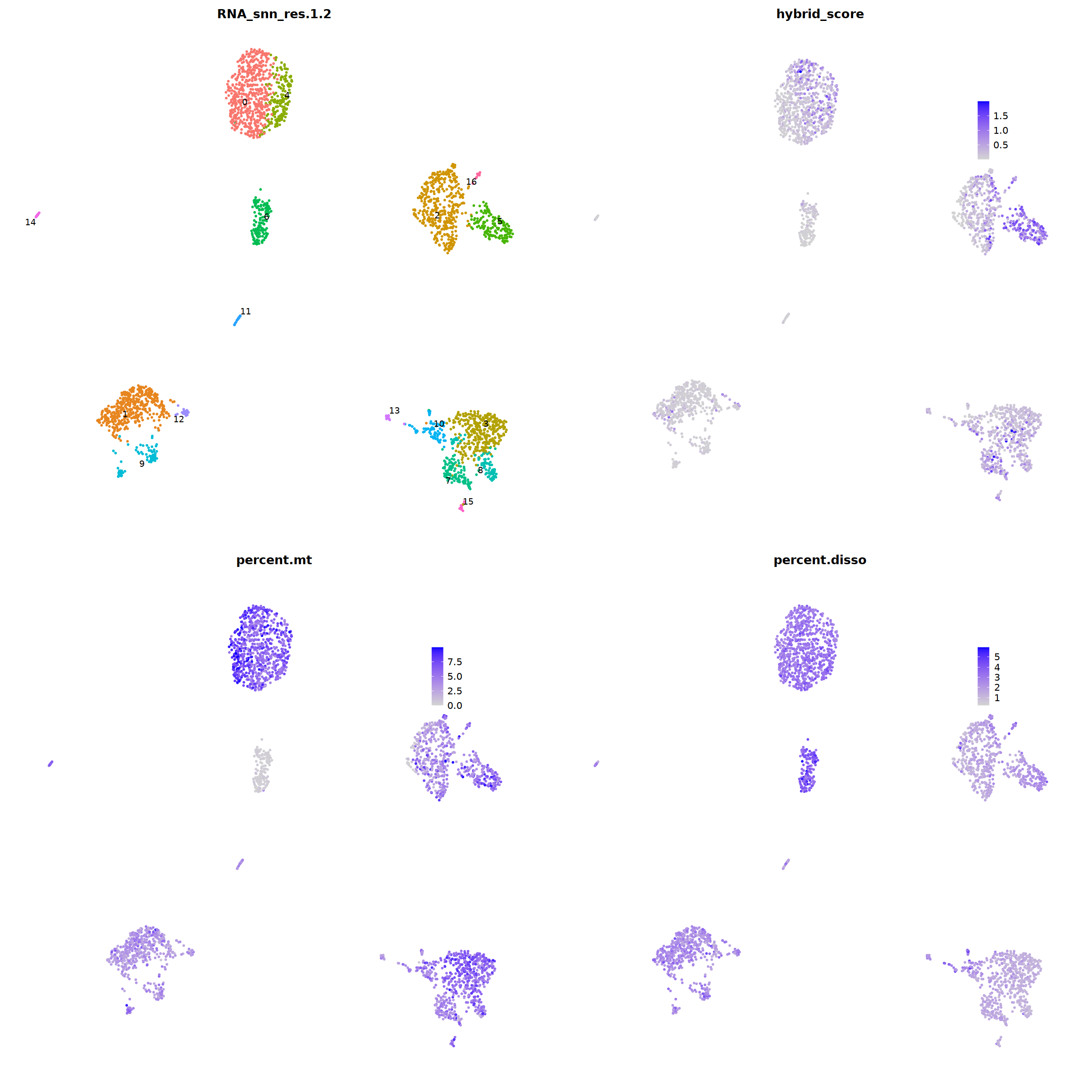
i=3
o(20,20)
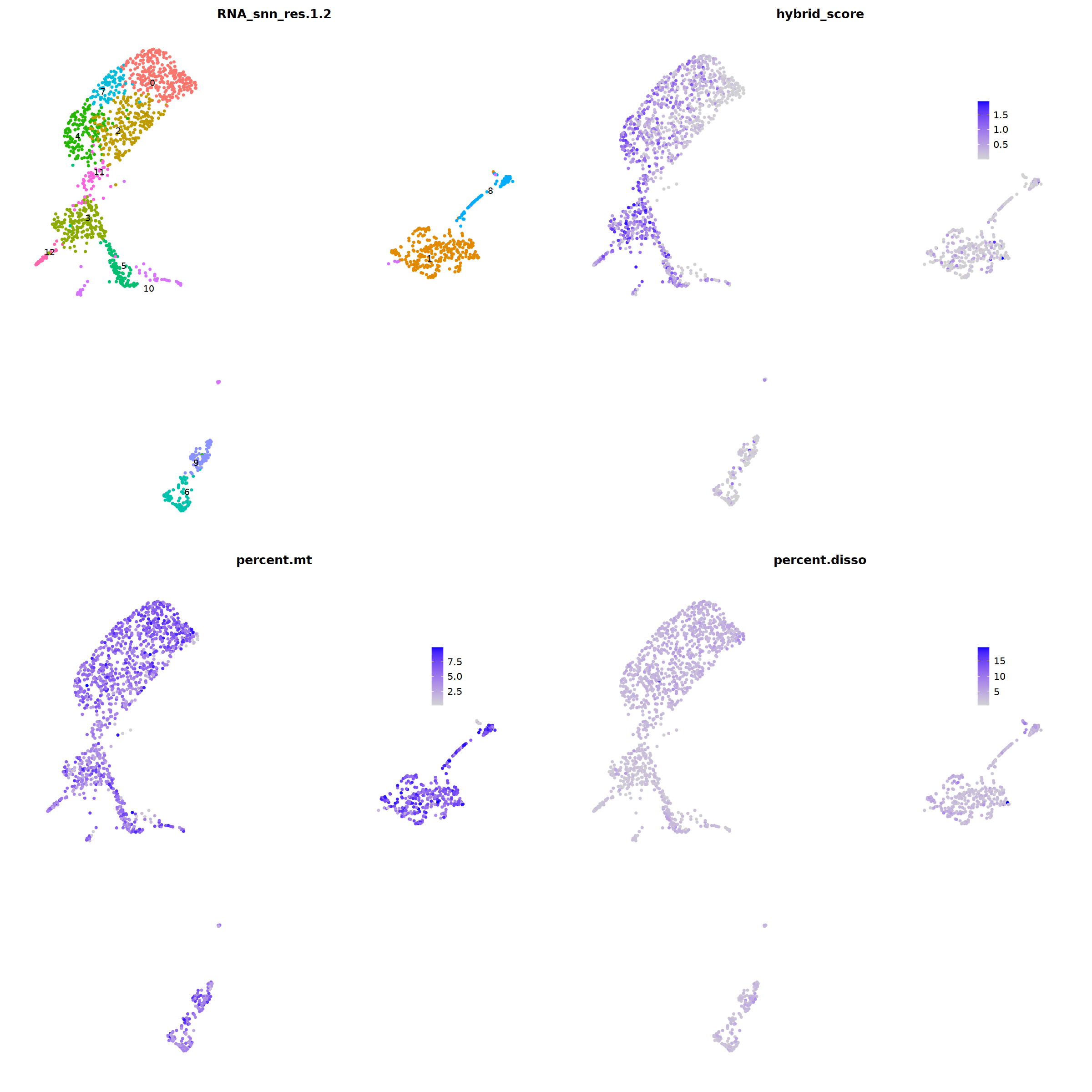
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
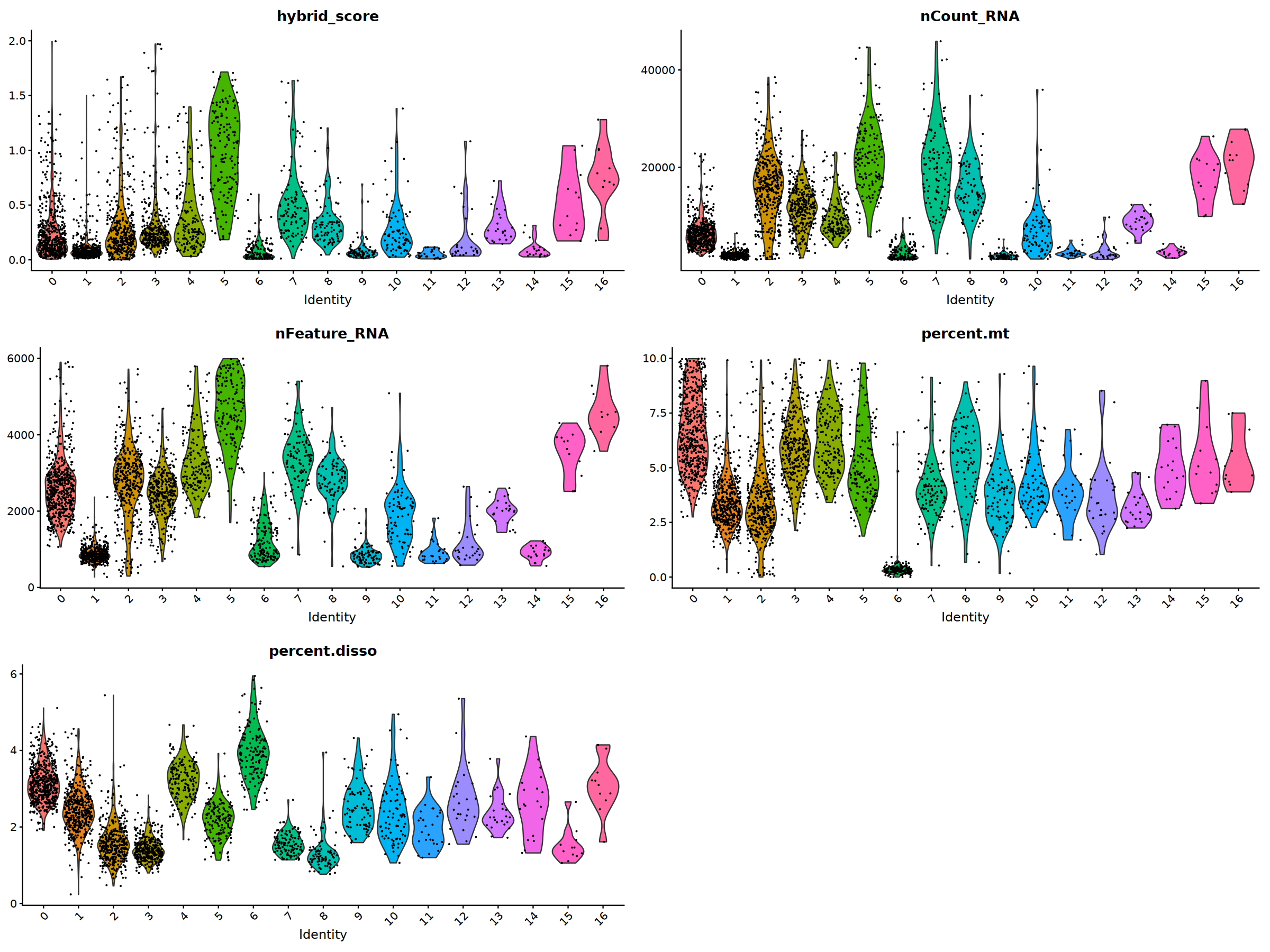
o(20,15)
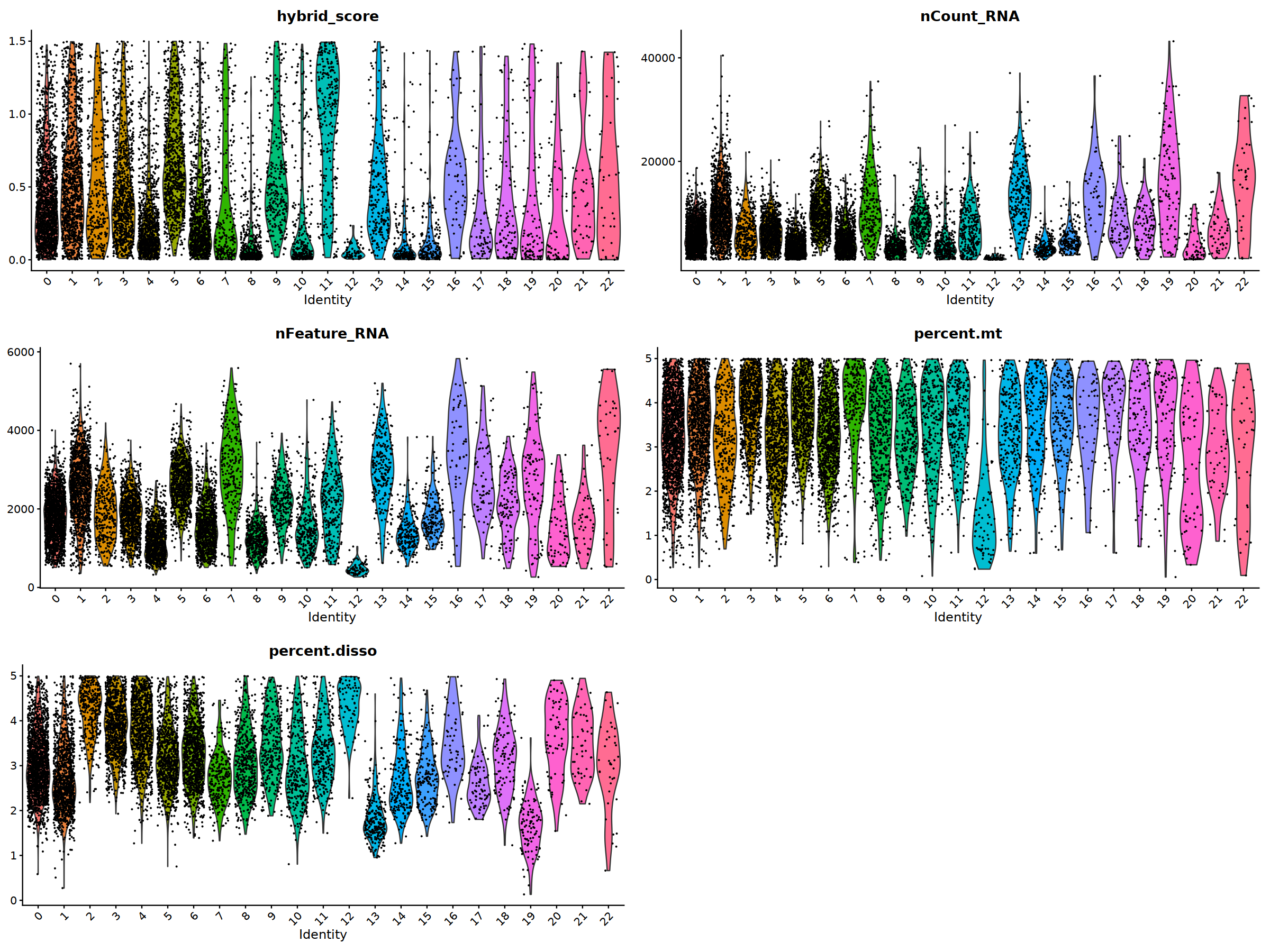
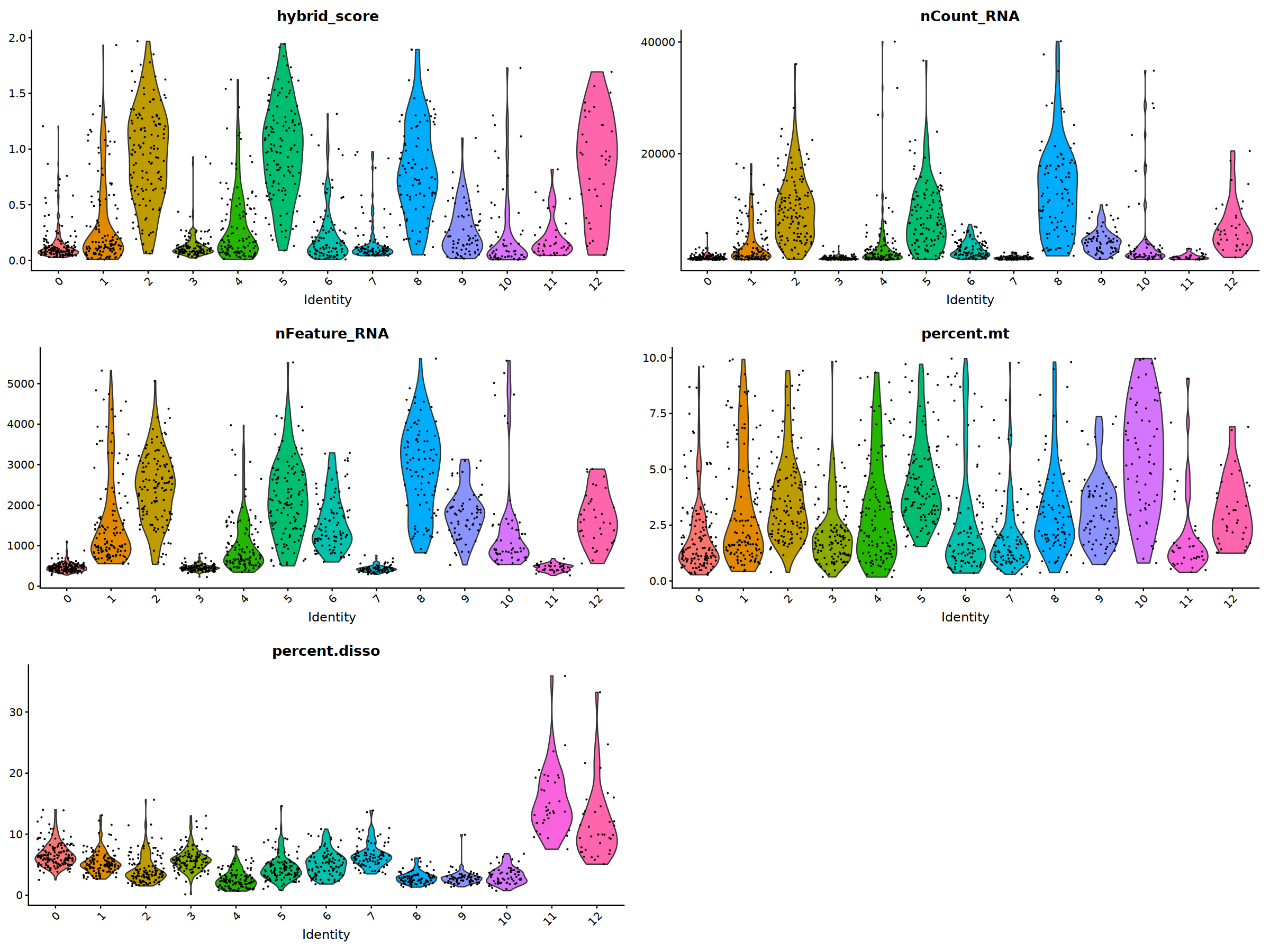
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
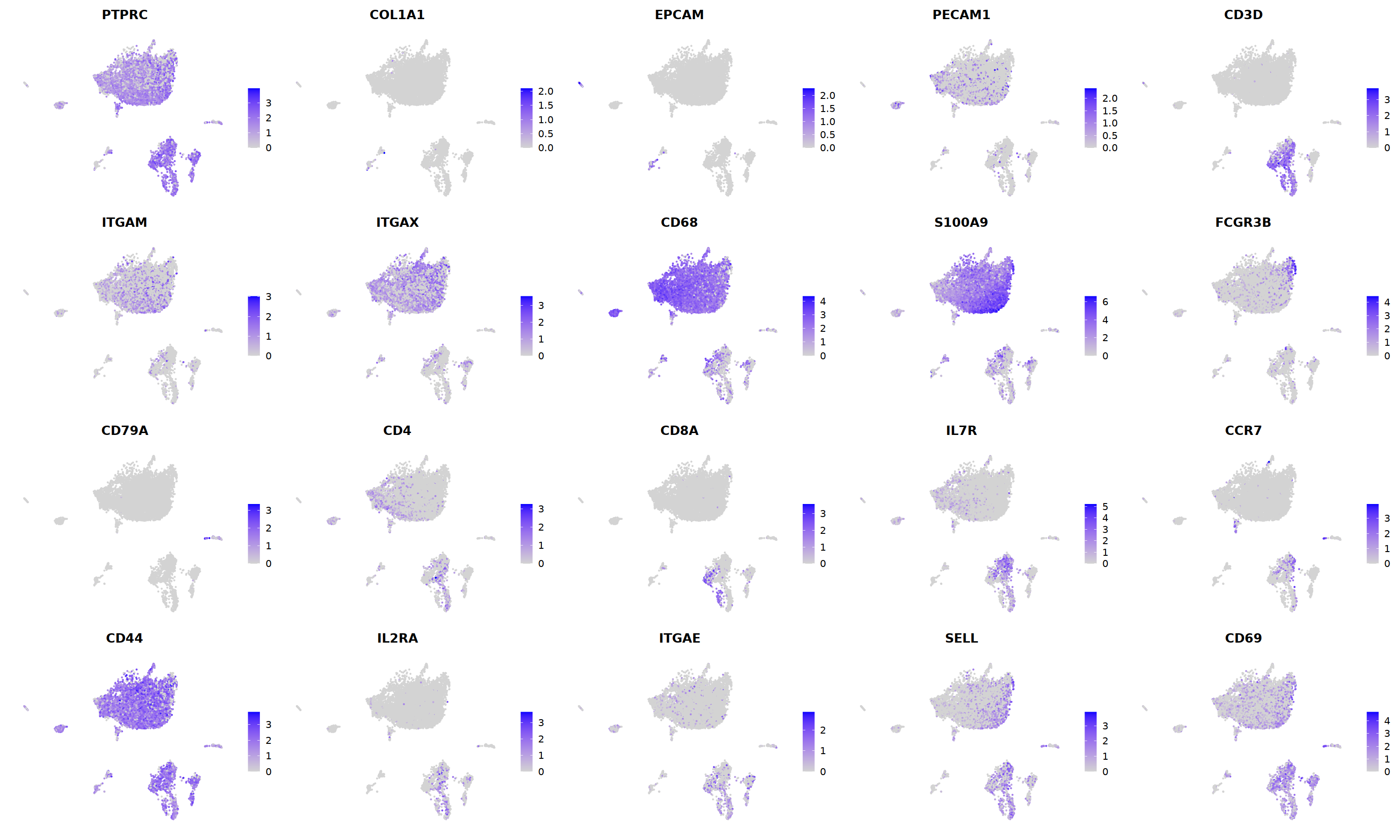
o(5*5, 5*3)
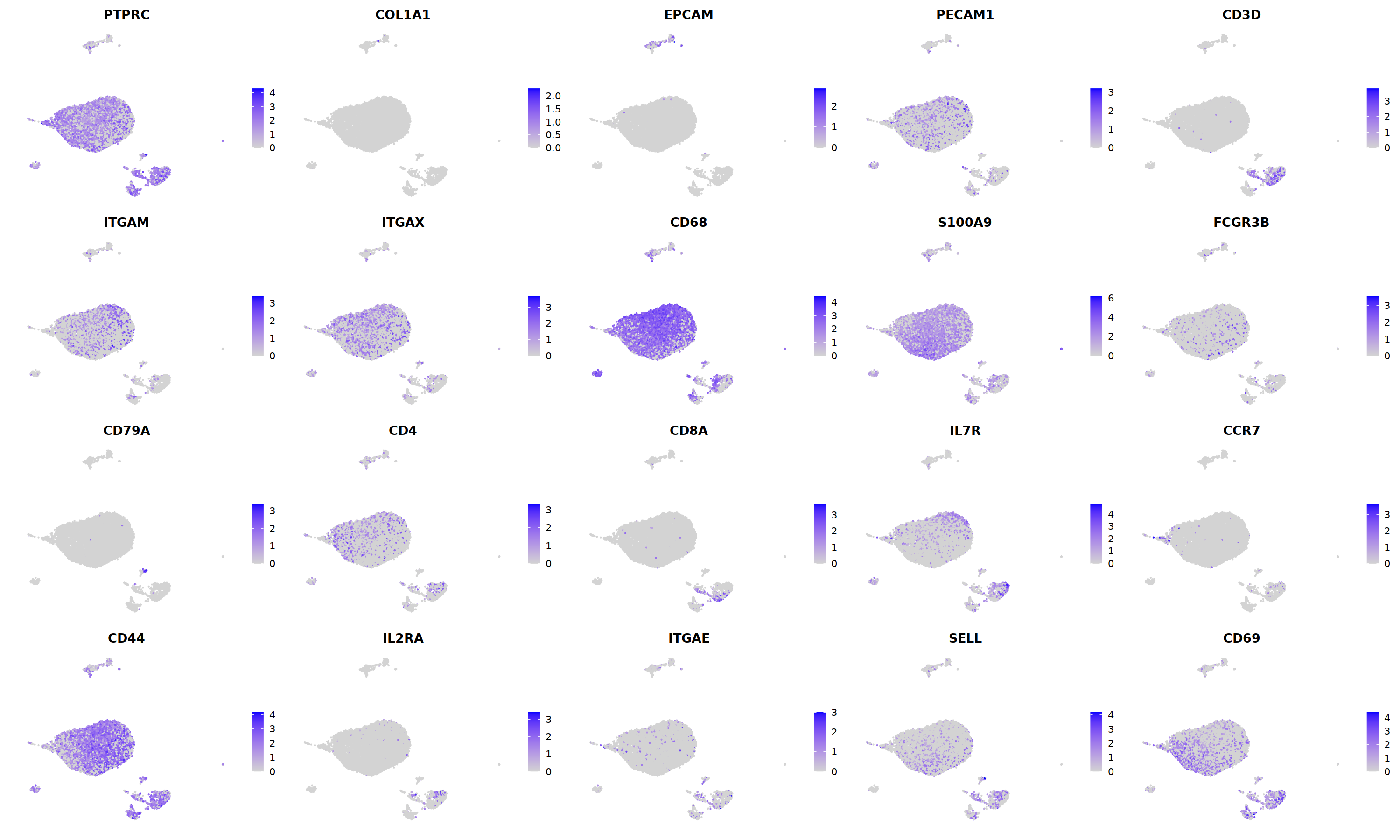
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
sample 4 (low quality)¶
i=4
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
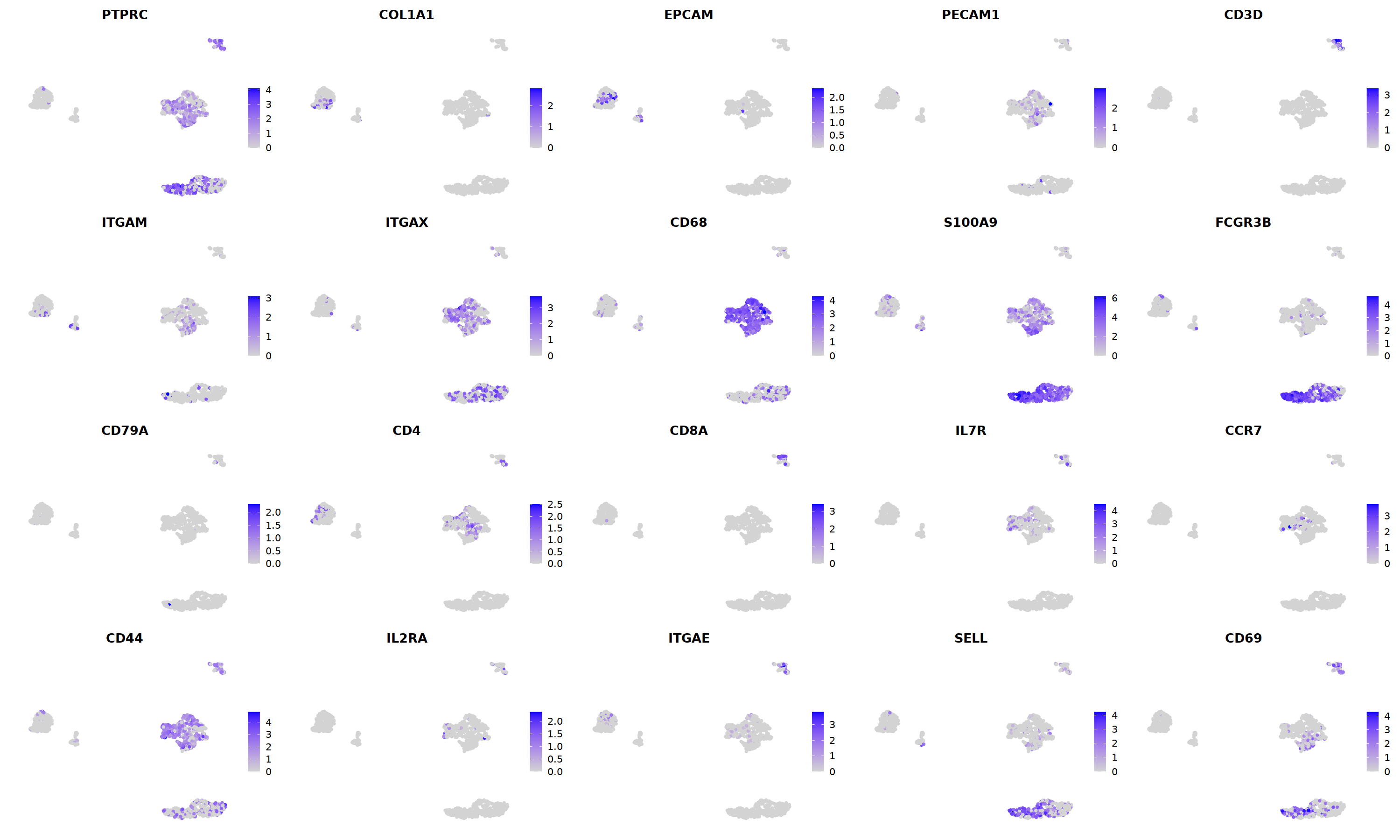
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.

Warning message in FeaturePlot(nCoV.list[[i]], features = c("PTPRC", "COL1A1", "EPCAM", :
“All cells have the same value (0) of COL1A1.”
sample 5¶
i=5
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nFeature_RNA < 3500)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nCount_RNA < 20000)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.75)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 7.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , RNA_snn_res.1.2 != 15)
sample 6¶
i=6
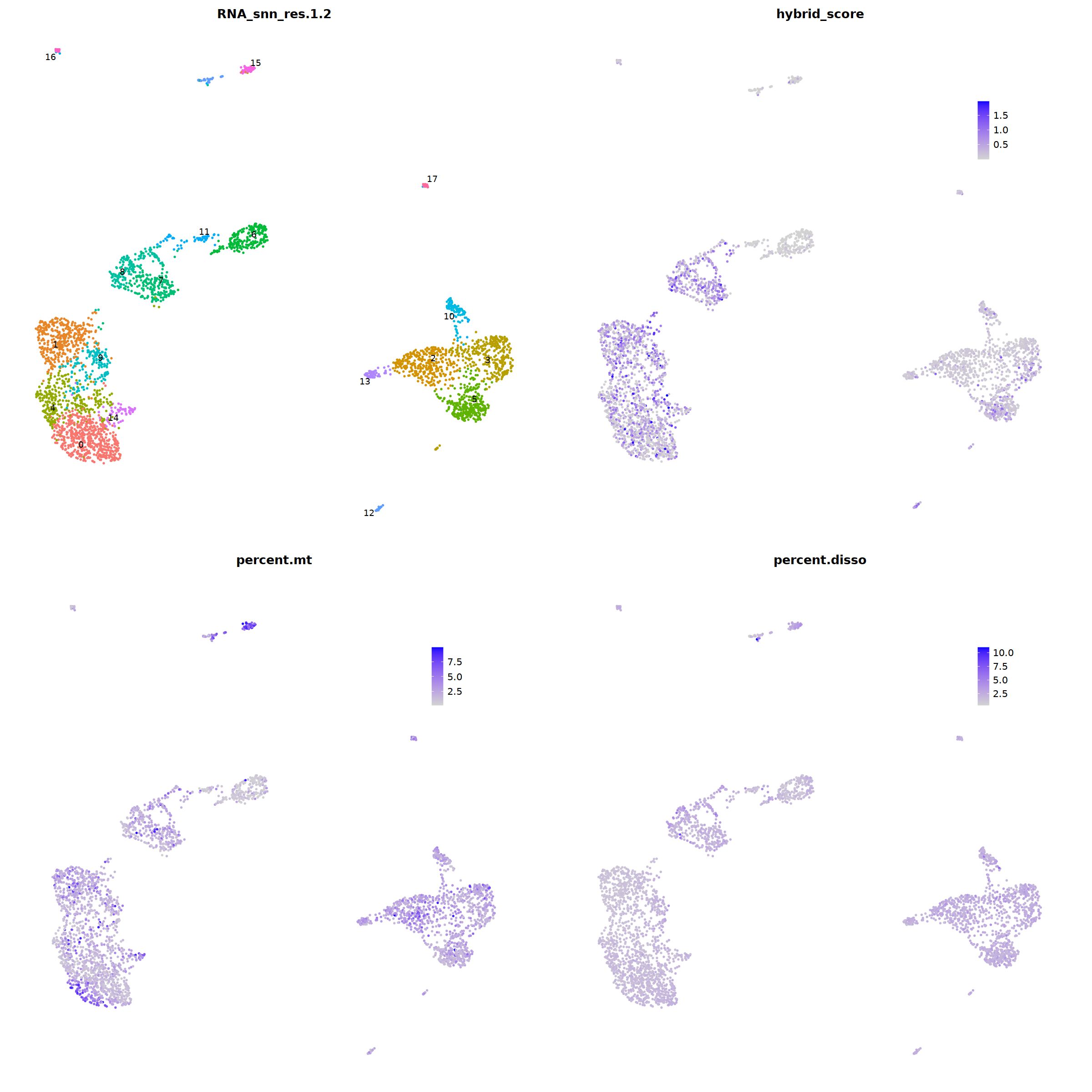
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.

nCoV.list[[i]] <- subset(nCoV.list[[i]] , nFeature_RNA < 4000)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nCount_RNA < 30000)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , RNA_snn_res.1.2 != 11)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , RNA_snn_res.1.2 != 12)
sample 7¶
i=7
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
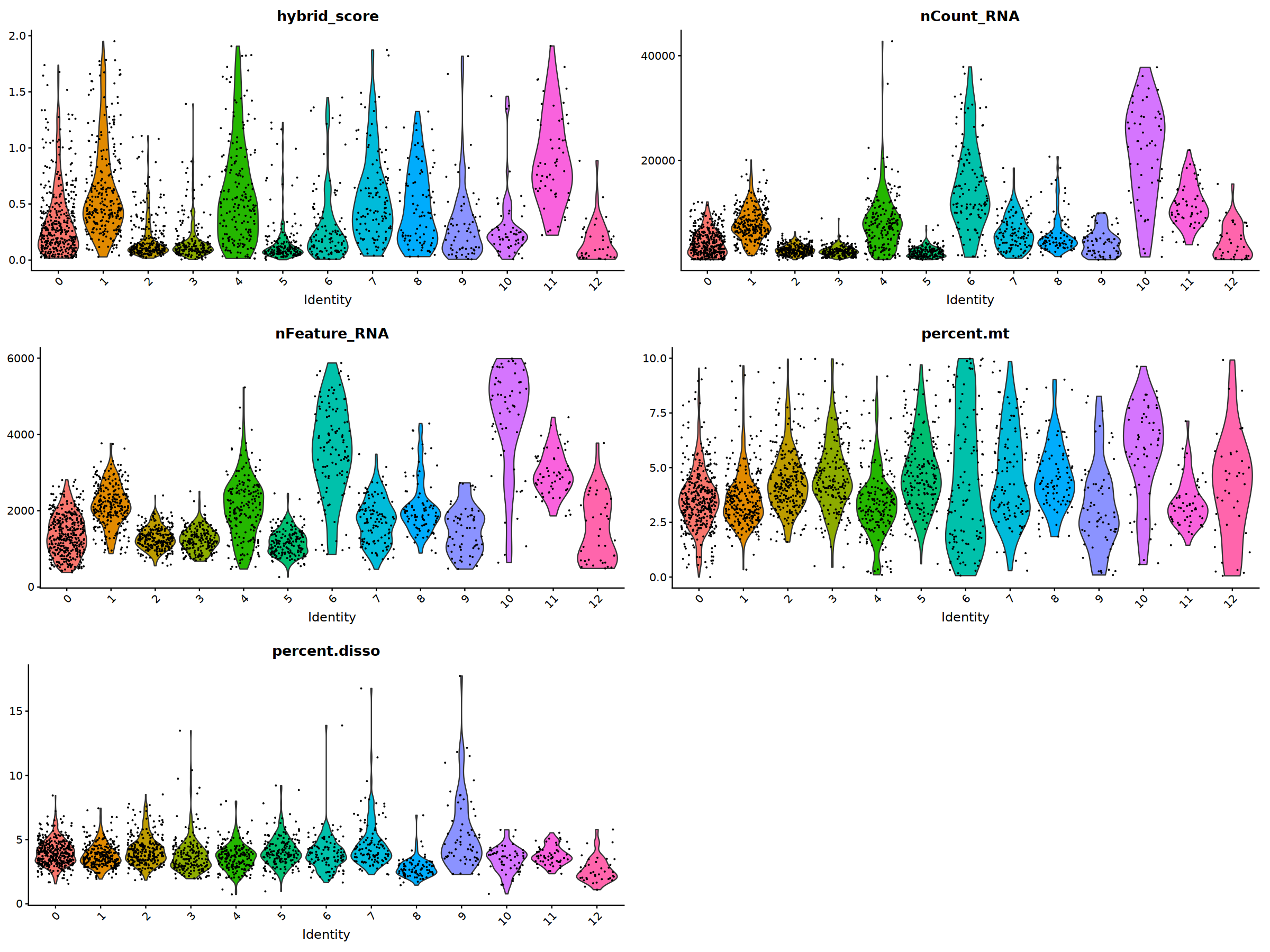
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso<10)
sample 8¶
i=8
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 10)
sample 9¶
i=9
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
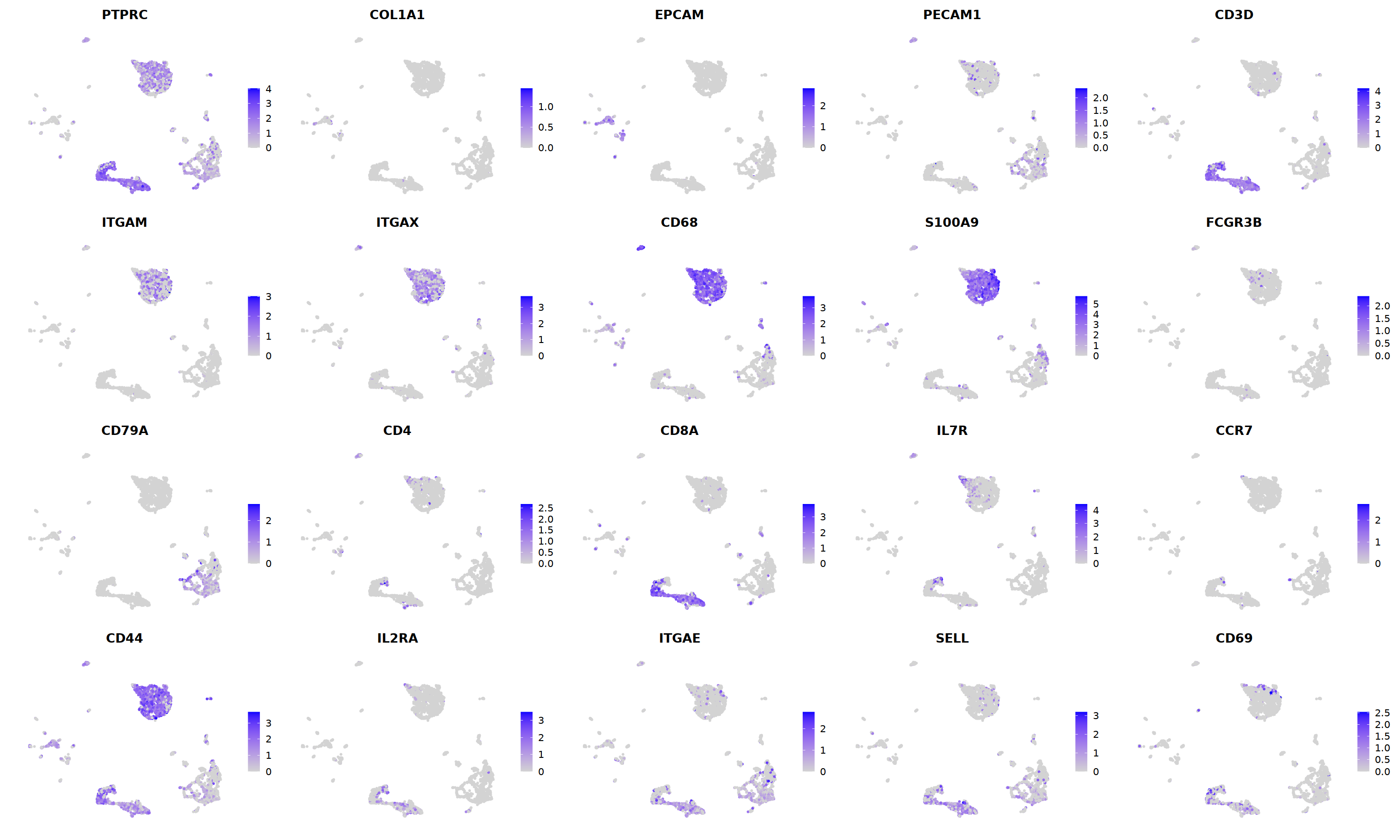
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5 )
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nFeature_RNA >500 )
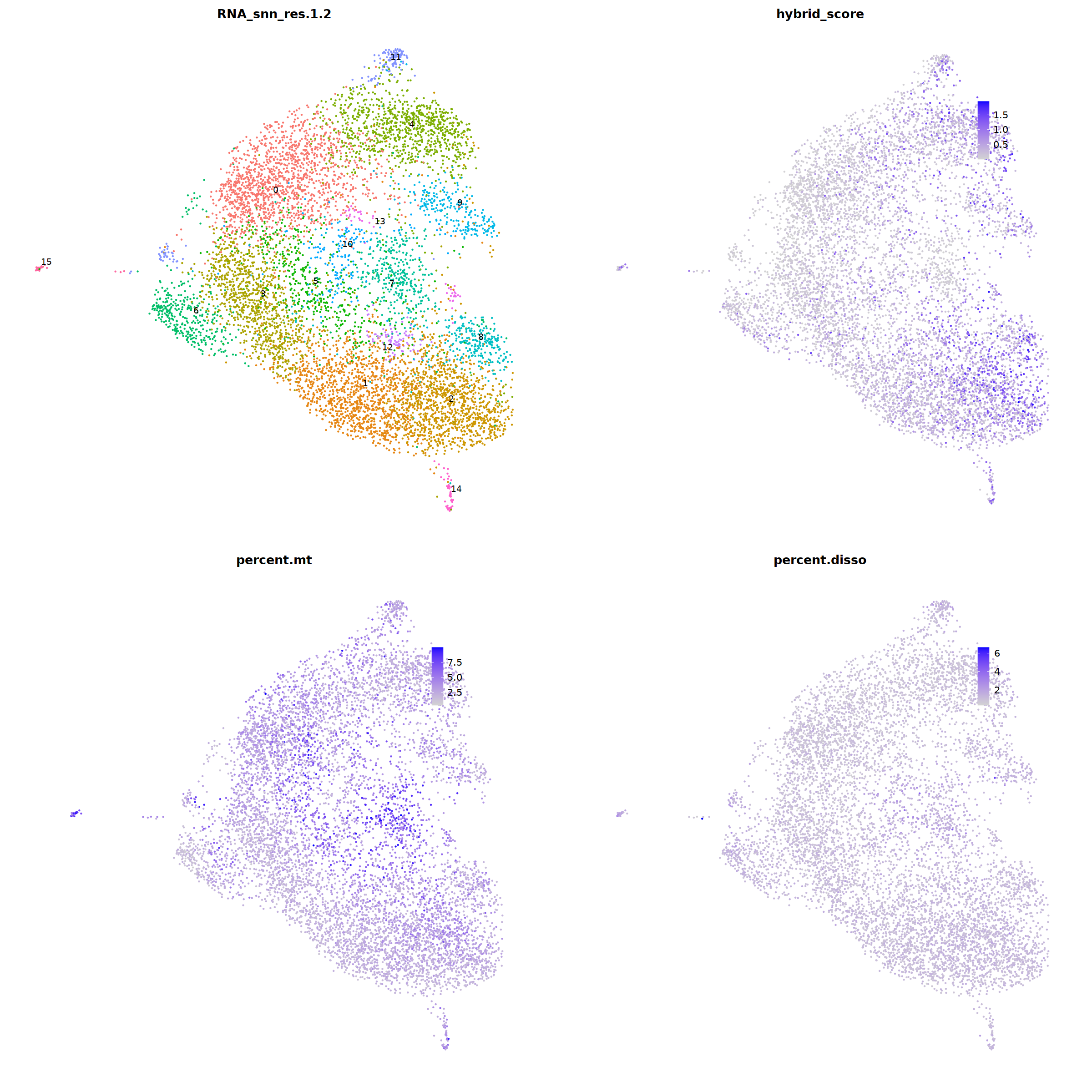
sample 10 (low quality)¶
i=10
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
Warning message in FeaturePlot(nCoV.list[[i]], features = c("PTPRC", "COL1A1", "EPCAM", :
“All cells have the same value (0) of COL1A1.”
sample 11 (low quality)¶
i=11
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
Warning message in FeaturePlot(nCoV.list[[i]], features = c("PTPRC", "COL1A1", "EPCAM", :
“All cells have the same value (0) of COL1A1.”
Warning message in FeaturePlot(nCoV.list[[i]], features = c("PTPRC", "COL1A1", "EPCAM", :
“All cells have the same value (0) of CD79A.”
sample 12¶
i=12
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
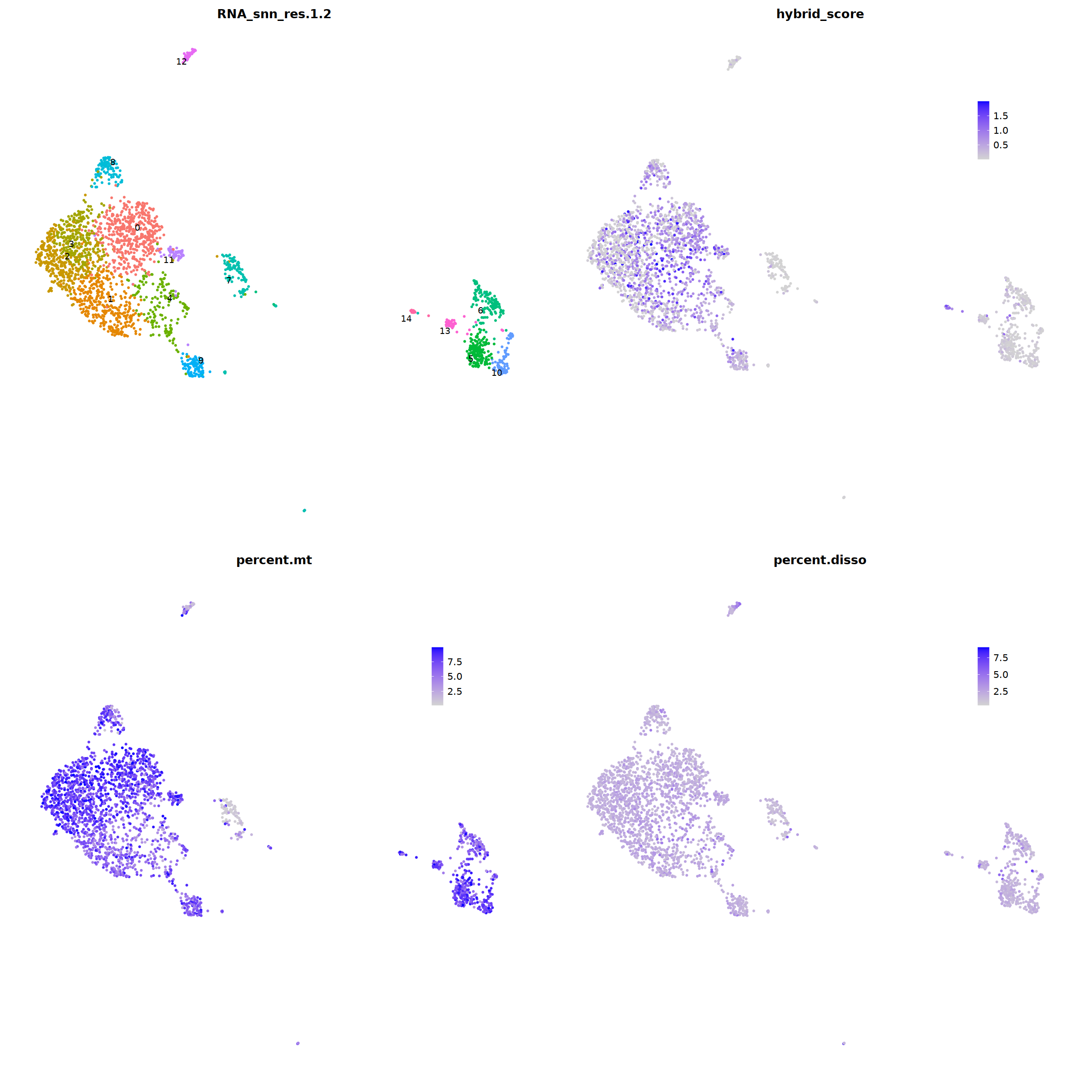
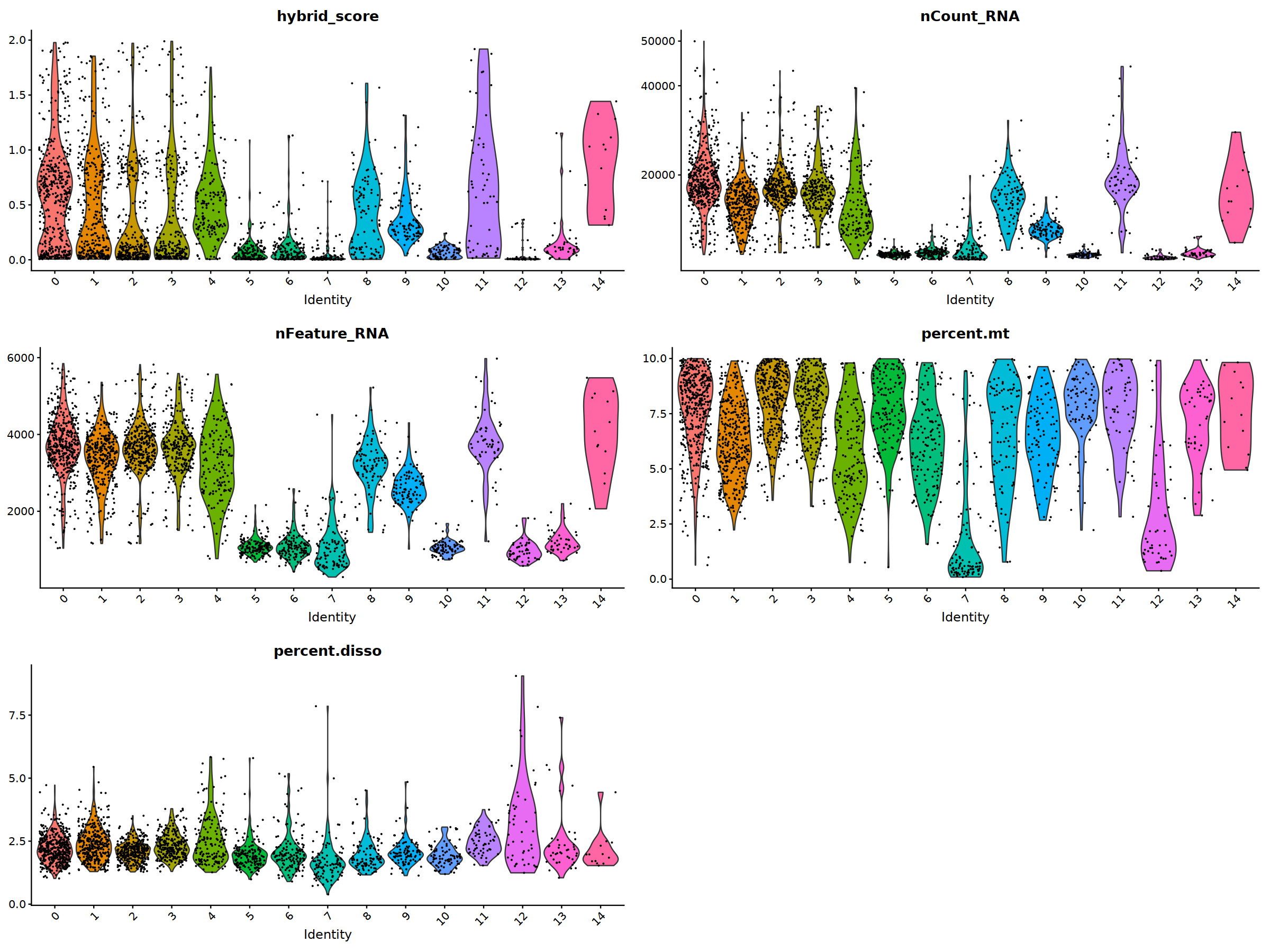
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nFeature_RNA < 5000)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , nCount_RNA < 40000)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5)
sample 13¶
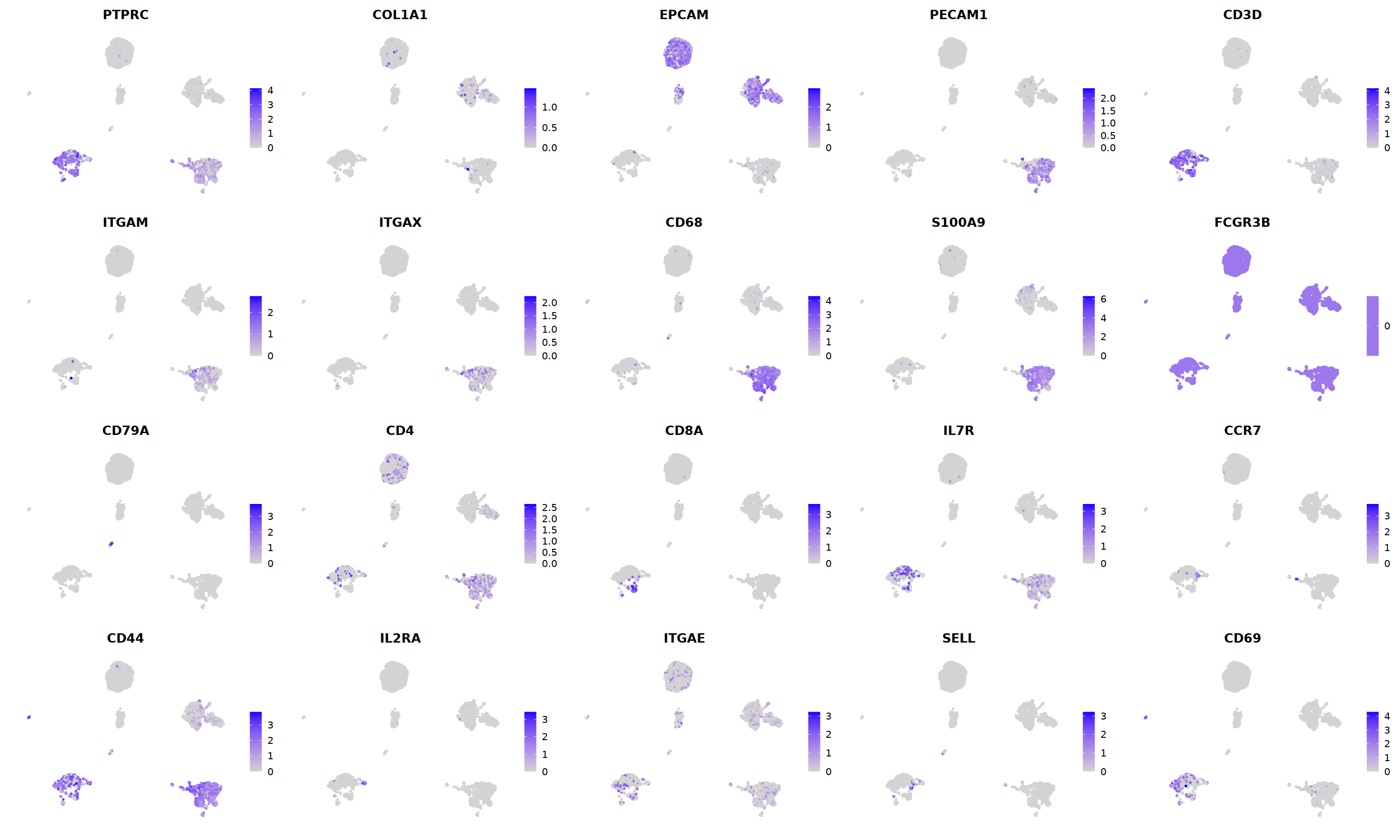
i=13
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
o(20,15)
ggarrange(
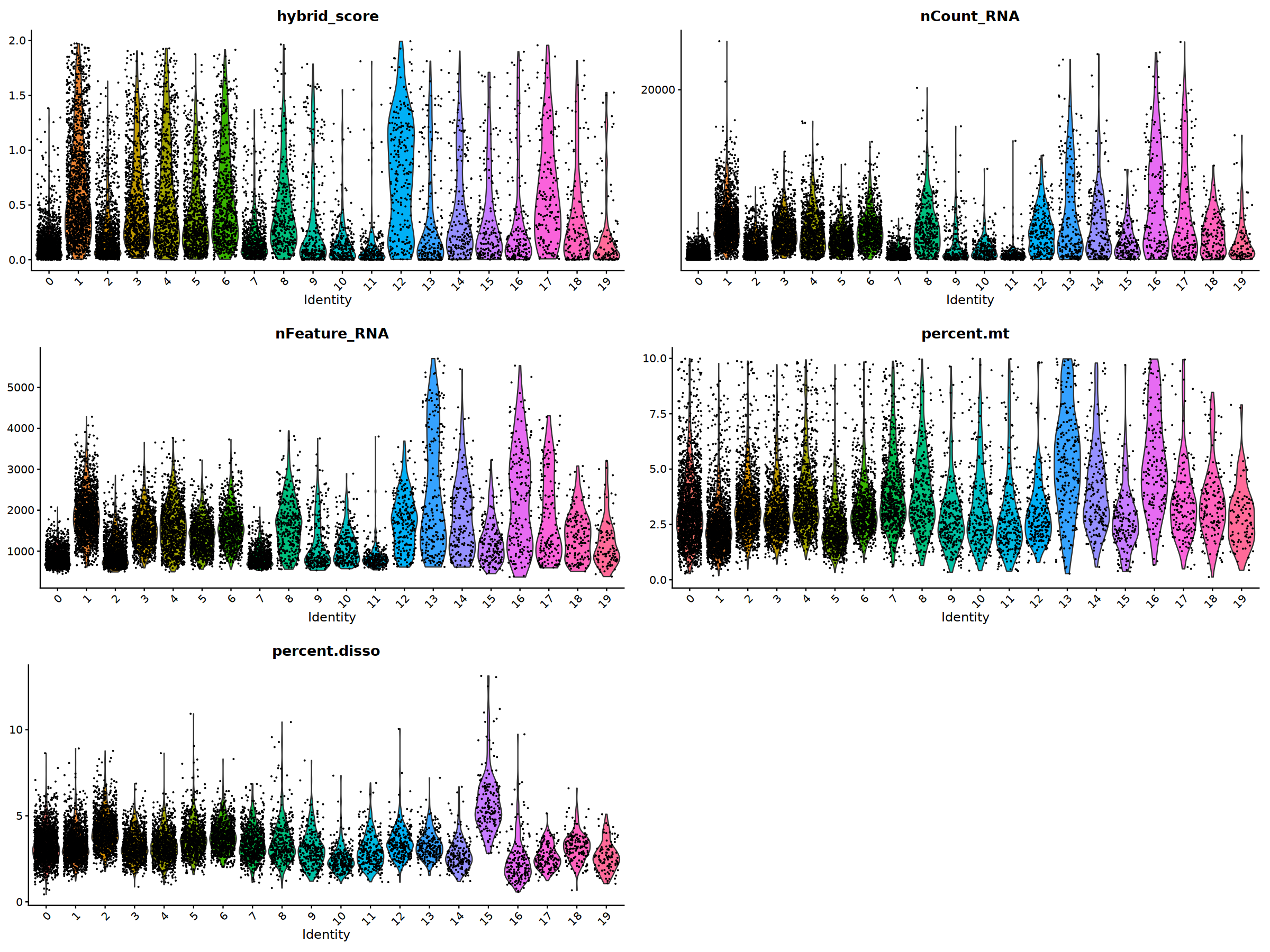
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend()&scale_y_continuous(breaks=c(20000,40000,50000,60000,70000,80000)),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)
o(5*5, 5*3)
FeaturePlot(nCoV.list[[i]],
features = c("PTPRC","COL1A1","EPCAM","PECAM1","CD3D",
'ITGAM','ITGAX',"CD68",'S100A9',"FCGR3B",
'CD79A','CD4','CD8A','IL7R','CCR7',
"CD44","IL2RA",'ITGAE','SELL','CD69'),
ncol=5
)&theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
)
Scale for 'y' is already present. Adding another scale for 'y', which will
replace the existing scale.
Warning message in FeaturePlot(nCoV.list[[i]], features = c("PTPRC", "COL1A1", "EPCAM", :
“All cells have the same value (0) of FCGR3B.”
nCoV.list[[i]] <- subset(nCoV.list[[i]] , hybrid_score < 1.5)
nCoV.list[[i]] <- subset(nCoV.list[[i]] , percent.disso < 5)
reclustering¶
library(scds)
dissociation.genes.hs<-c("ACTG1","ANKRD1","ARID5A","ATF3","ATF4","BAG3","BHLHE40","CCNL1","CCRN4L",
"CEBPB","CEBPD","CEBPG","CSRNP1","CXCL1","CYR61","DCN","DDX3XX","DDX5","DES","DNAJA1","DNAJB1",
"DNAJB4","DUSP1","DUSP8","EGR1","EGR2","EIF1","EIF5","ERF","ERRFI1","FAM132B","FOS","FOSB","FOSL2",
"GADD45A","GADD45G","BRD2","BTG1","BTG2","GCC1","GEM","H3F3B","HIPK3","HSP90AA1","HSP90AB1",
"HSPA1A","HSPA1B","HSPA5","HSPA8","HSPB1","HSPE1","HSPH1","ID3","IDI1","IER2","IER3","IER5",
"IFRD1","IL6","IRF1","IRF8","ITPKC","JUN","JUNB","JUND","KCNE4","KLF2","KLF4","KLF6","KLF9",
"LITAF","LMNA","MAFF","MAFK","MCL1","MIDN","MIR22HG","MT1","MT2","MYADM","MYC","MYD88","NCKAP5L",
"NCOA7","NFKBIA","NFKBIZ","NOP58","NPPC","NR4A1","ODC1","OSGIN1","OXNAD1","PCF11","PDE4B","PER1",
"PHLDA1","PNP","PNRC1","PPP1CC","PPP1R15A","PXDC1","RAP1B","RASSF1","RHOB","RHOH","RIPK1","SAT1X",
"SBNO2","SDC4","SERPINE1","SKIL","SLC10A6","SLC38A2","SLC41A1","SOCS3","SQSTM1","SRF","SRSF5",
"SRSF7","STAT3","TAGLN2","TIPARP","TNFAIP3","TNFAIP6","TPM3","TPPP3","TRA2A","TRA2B","TRIB1",
"TUBB4B","TUBB6","UBC","USP2","WAC","ZC3H12A","ZFAND5","ZFP36","ZFP36L1","ZFP36L2","ZYX")
# normalize and identify variable features for each dataset independently
nCoV.list <- lapply(X = nCoV.list, FUN = function(x) {
DefaultAssay(x)<-'RNA'
x <- NormalizeData(x, verbose = F)
x <- FindVariableFeatures(x, selection.method = "vst", nfeatures = 2000, verbose = F)
x <- ScaleData(x, features = rownames(x), verbose = F)
x <- RunPCA(object = x, features = VariableFeatures(x), npcs = 50, verbose = F)
x <- FindNeighbors(x, reduction = "pca", dims = 1:30, verbose = F)
x <- RunUMAP(object=x,reduction = "pca", dims = 1:30, verbose = F)
x <- FindClusters(object=x, resolution = c(0.7,0.9,1.2),verbose = F)
(x %>%
as.SingleCellExperiment %>%
cxds_bcds_hybrid)@colData[,c('cxds_score','bcds_score','hybrid_score')
] %>% as.data.frame -> scds.doublet.profiles
meta <- merge(x@meta.data, scds.doublet.profiles, by.x=0, by.y=0)
rownames(meta) <- meta$Row.names
meta$Row.names <- NULL
x@meta.data <- meta
gset <- dissociation.genes.hs
gset <- gset[gset %in% rownames(x)]
x[["percent.disso"]]<-PercentageFeatureSet(x, features = gset)
x
})
library(tictoc)
tic()
save(nCoV.list,
file = 'nCoV.list.qc.rda',
compress = T, compression_level = 9)
toc()
2695.756 sec elapsed