read data¶
o<-function(w,h) options(repr.plot.width=w, repr.plot.height=h)
C141 <- Read10X_h5("covid/GSM4339769_C141_filtered_feature_bc_matrix.h5")
C142 <- Read10X_h5("covid/GSM4339770_C142_filtered_feature_bc_matrix.h5")
C143 <- Read10X_h5("covid/GSM4339771_C143_filtered_feature_bc_matrix.h5")
C144 <- Read10X_h5("covid/GSM4339772_C144_filtered_feature_bc_matrix.h5")
C145 <- Read10X_h5("covid/GSM4339773_C145_filtered_feature_bc_matrix.h5")
C146 <- Read10X_h5("covid/GSM4339774_C146_filtered_feature_bc_matrix.h5")
C148 <- Read10X_h5("covid/GSM4475051_C148_filtered_feature_bc_matrix.h5")
C149 <- Read10X_h5("covid/GSM4475052_C149_filtered_feature_bc_matrix.h5")
C152 <- Read10X_h5("covid/GSM4475053_C152_filtered_feature_bc_matrix.h5")
C51 <- Read10X_h5("covid/GSM4475048_C51_filtered_feature_bc_matrix.h5" )
C52 <- Read10X_h5("covid/GSM4475049_C52_filtered_feature_bc_matrix.h5" )
C100 <- Read10X_h5("covid/GSM4475050_C100_filtered_feature_bc_matrix.h5")
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
Warning message in sparseMatrix(i = indices[] + 1, p = indptr[], x = as.numeric(x = counts[]), :
“'giveCsparse' has been deprecated; setting 'repr = "T"' for you”
GSM3660650<- Read10X(data.dir = 'covid/GSM3660650_SC249NORbal_fresh')
C141<-CreateSeuratObject(counts = C141, project = "C141",min.cells = 3, min.features = 200)
C142<-CreateSeuratObject(counts = C142, project = "C142",min.cells = 3, min.features = 200)
C143<-CreateSeuratObject(counts = C143, project = "C143",min.cells = 3, min.features = 200)
C144<-CreateSeuratObject(counts = C144, project = "C144",min.cells = 3, min.features = 200)
C145<-CreateSeuratObject(counts = C145, project = "C145",min.cells = 3, min.features = 200)
C146<-CreateSeuratObject(counts = C146, project = "C146",min.cells = 3, min.features = 200)
C148<-CreateSeuratObject(counts = C148, project = "C148",min.cells = 3, min.features = 200)
C149<-CreateSeuratObject(counts = C149, project = "C149",min.cells = 3, min.features = 200)
C152<-CreateSeuratObject(counts = C152, project = "C152",min.cells = 3, min.features = 200)
C51<-CreateSeuratObject(counts = C51, project = "C51",min.cells = 3, min.features = 200)
C52<-CreateSeuratObject(counts = C52, project = "C52",min.cells = 3, min.features = 200)
C100<-CreateSeuratObject(counts = C100, project = "C100",min.cells = 3, min.features = 200)
GSM3660650<-CreateSeuratObject(counts = GSM3660650, project = "GSM3660650",min.cells = 3, min.features = 200)
C141$group<-"mild"
C142$group<-"mild"
C143$group<-"severe"
C144$group<-"mild"
C145$group<-"severe"
C146$group<-"severe"
C148$group<-"severe"
C149$group<-"severe"
C152$group<-"severe"
C51$group <- "healthy"
C52$group <- "healthy"
C100$group <- "healthy"
GSM3660650$group <- "healthy"
# We first calculate mt-gene fractions and visualize them
C141[["percent.mt"]]<-PercentageFeatureSet(C141,pattern = "^MT")
p141<-VlnPlot(C141, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C142[["percent.mt"]]<-PercentageFeatureSet(C142,pattern = "^MT")
p142<-VlnPlot(C142, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C143[["percent.mt"]]<-PercentageFeatureSet(C143,pattern = "^MT")
p143<-VlnPlot(C143, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C144[["percent.mt"]]<-PercentageFeatureSet(C144,pattern = "^MT")
p144<-VlnPlot(C144, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C145[["percent.mt"]]<-PercentageFeatureSet(C145,pattern = "^MT")
p145<-VlnPlot(C145, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C146[["percent.mt"]]<-PercentageFeatureSet(C146,pattern = "^MT")
p146<-VlnPlot(C146, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C148[["percent.mt"]]<-PercentageFeatureSet(C148,pattern = "^MT")
p148<-VlnPlot(C148, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C149[["percent.mt"]]<-PercentageFeatureSet(C149,pattern = "^MT")
p149<-VlnPlot(C149, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C152[["percent.mt"]]<-PercentageFeatureSet(C152,pattern = "^MT")
p152<-VlnPlot(C152, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C51[["percent.mt"]]<-PercentageFeatureSet(C51,pattern = "^MT")
p51<-VlnPlot(C51, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C52[["percent.mt"]]<-PercentageFeatureSet(C52,pattern = "^MT")
p52<-VlnPlot(C52, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C100[["percent.mt"]]<-PercentageFeatureSet(C100,pattern = "^MT")
p100<-VlnPlot(C100, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
GSM3660650[["percent.mt"]]<-PercentageFeatureSet(GSM3660650,pattern = "^MT")
pGSM3660650<-VlnPlot(GSM3660650, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
C141 <- subset(C141, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C142 <- subset(C142, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C143 <- subset(C143, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C144 <- subset(C144, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C145 <- subset(C145, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C146 <- subset(C146, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C148 <- subset(C148, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C149 <- subset(C149, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C152 <- subset(C152, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C51 <- subset(C51, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C52 <- subset(C52, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C100 <- subset(C100, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
GSM3660650 <- subset(GSM3660650, subset = nFeature_RNA > 200 & nFeature_RNA < 6000 & percent.mt < 10 & nCount_RNA >1000)
C141 <- NormalizeData(C141,verbose = F)
C142 <- NormalizeData(C142,verbose = F)
C143 <- NormalizeData(C143,verbose = F)
C144 <- NormalizeData(C144,verbose = F)
C145 <- NormalizeData(C145,verbose = F)
C146 <- NormalizeData(C146,verbose = F)
C148 <- NormalizeData(C148,verbose = F)
C149 <- NormalizeData(C149,verbose = F)
C152 <- NormalizeData(C152,verbose = F)
C51 <- NormalizeData(C51,verbose = F)
C52 <- NormalizeData(C52,verbose = F)
C100 <- NormalizeData(C100,verbose = F)
GSM3660650<- NormalizeData(GSM3660650,verbose = F)
C141 <- FindVariableFeatures(C141, selection.method = "vst", nfeatures = 2000)
C142 <- FindVariableFeatures(C142, selection.method = "vst", nfeatures = 2000)
C143 <- FindVariableFeatures(C143, selection.method = "vst", nfeatures = 2000)
C144 <- FindVariableFeatures(C144, selection.method = "vst", nfeatures = 2000)
C145 <- FindVariableFeatures(C145, selection.method = "vst", nfeatures = 2000)
C146 <- FindVariableFeatures(C146, selection.method = "vst", nfeatures = 2000)
C148 <- FindVariableFeatures(C148, selection.method = "vst", nfeatures = 2000)
C149 <- FindVariableFeatures(C149, selection.method = "vst", nfeatures = 2000)
C152 <- FindVariableFeatures(C152, selection.method = "vst", nfeatures = 2000)
C51 <- FindVariableFeatures(C51, selection.method = "vst", nfeatures = 2000)
C52 <- FindVariableFeatures(C52, selection.method = "vst", nfeatures = 2000)
C100 <- FindVariableFeatures(C100, selection.method = "vst", nfeatures = 2000)
GSM3660650 <- FindVariableFeatures(GSM3660650, selection.method = "vst", nfeatures = 2000)
integrate data¶
nCoV.list <- list(C141= C141,C142= C142,C143= C143,C144= C144,C145= C145,
C146= C146,C148= C148,C149= C149,C152= C152,C51= C51 ,
C52= C52 ,C100= C100,GSM3660650= GSM3660650)
# select features that are repeatedly variable across datasets for integration
features <- SelectIntegrationFeatures(object.list = nCoV.list)
library(future)
options(future.globals.maxSize = 1000 * 1024^2*5)
plan("multiprocess", workers = 15)
nCoV.anchors <- FindIntegrationAnchors(object.list = nCoV.list, anchor.features = features)
nCoV.integrated <- IntegrateData(anchorset = nCoV.anchors)
plan("sequential")
Warning message in CheckDuplicateCellNames(object.list = object.list):
“Some cell names are duplicated across objects provided. Renaming to enforce unique cell names.”
Scaling features for provided objects
Finding all pairwise anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8807 anchors
Filtering anchors
Retained 6635 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 12279 anchors
Filtering anchors
Retained 2232 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 11752 anchors
Filtering anchors
Retained 2246 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1584 anchors
Filtering anchors
Retained 1546 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1595 anchors
Filtering anchors
Retained 1561 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1658 anchors
Filtering anchors
Retained 1535 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 11271 anchors
Filtering anchors
Retained 2583 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 10846 anchors
Filtering anchors
Retained 2206 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 33359 anchors
Filtering anchors
Retained 9840 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1585 anchors
Filtering anchors
Retained 629 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4210 anchors
Filtering anchors
Retained 2074 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4115 anchors
Filtering anchors
Retained 2069 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4951 anchors
Filtering anchors
Retained 3518 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1367 anchors
Filtering anchors
Retained 890 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4870 anchors
Filtering anchors
Retained 2607 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5118 anchors
Filtering anchors
Retained 2922 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4912 anchors
Filtering anchors
Retained 2575 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6555 anchors
Filtering anchors
Retained 5164 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1472 anchors
Filtering anchors
Retained 1112 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6364 anchors
Filtering anchors
Retained 4490 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 3720 anchors
Filtering anchors
Retained 2889 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6409 anchors
Filtering anchors
Retained 3343 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6419 anchors
Filtering anchors
Retained 3564 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7636 anchors
Filtering anchors
Retained 5139 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1609 anchors
Filtering anchors
Retained 1001 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6964 anchors
Filtering anchors
Retained 4282 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 3599 anchors
Filtering anchors
Retained 2338 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4555 anchors
Filtering anchors
Retained 3072 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6442 anchors
Filtering anchors
Retained 2840 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6395 anchors
Filtering anchors
Retained 2749 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8831 anchors
Filtering anchors
Retained 4652 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1633 anchors
Filtering anchors
Retained 916 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8416 anchors
Filtering anchors
Retained 3887 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 3974 anchors
Filtering anchors
Retained 2263 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4680 anchors
Filtering anchors
Retained 2964 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5594 anchors
Filtering anchors
Retained 3043 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 10428 anchors
Filtering anchors
Retained 2724 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 10222 anchors
Filtering anchors
Retained 2722 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 26281 anchors
Filtering anchors
Retained 3159 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1682 anchors
Filtering anchors
Retained 455 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 25872 anchors
Filtering anchors
Retained 2827 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4951 anchors
Filtering anchors
Retained 805 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6346 anchors
Filtering anchors
Retained 1259 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7227 anchors
Filtering anchors
Retained 1167 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8765 anchors
Filtering anchors
Retained 1417 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 11269 anchors
Filtering anchors
Retained 2164 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 11197 anchors
Filtering anchors
Retained 2083 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 27092 anchors
Filtering anchors
Retained 3126 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1605 anchors
Filtering anchors
Retained 319 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 26615 anchors
Filtering anchors
Retained 2740 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4855 anchors
Filtering anchors
Retained 947 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6030 anchors
Filtering anchors
Retained 1143 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7753 anchors
Filtering anchors
Retained 1006 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8959 anchors
Filtering anchors
Retained 1304 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 22573 anchors
Filtering anchors
Retained 4484 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7042 anchors
Filtering anchors
Retained 3989 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7010 anchors
Filtering anchors
Retained 3650 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 10122 anchors
Filtering anchors
Retained 4106 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1565 anchors
Filtering anchors
Retained 846 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 9744 anchors
Filtering anchors
Retained 3135 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4356 anchors
Filtering anchors
Retained 1678 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5041 anchors
Filtering anchors
Retained 2105 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5484 anchors
Filtering anchors
Retained 2553 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6079 anchors
Filtering anchors
Retained 1856 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8863 anchors
Filtering anchors
Retained 3774 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 9113 anchors
Filtering anchors
Retained 3259 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5410 anchors
Filtering anchors
Retained 3152 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5438 anchors
Filtering anchors
Retained 3219 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8872 anchors
Filtering anchors
Retained 3356 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 1645 anchors
Filtering anchors
Retained 1035 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 7739 anchors
Filtering anchors
Retained 2961 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4115 anchors
Filtering anchors
Retained 1664 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4290 anchors
Filtering anchors
Retained 2500 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 4821 anchors
Filtering anchors
Retained 2740 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 5640 anchors
Filtering anchors
Retained 2751 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 8822 anchors
Filtering anchors
Retained 1952 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 9351 anchors
Filtering anchors
Retained 1664 anchors
Running CCA
Merging objects
Finding neighborhoods
Finding anchors
Found 6050 anchors
Filtering anchors
Retained 2442 anchors
Error in slot(object = anchorset, name = "reference.objects"): object 'nCoV' not found
Traceback:
1. IntegrateData(anchorset = nCoV)
2. slot(object = anchorset, name = "reference.objects")
plan("sequential")
options(future.globals.maxSize = 1000 * 1024^2*5)
plan("multiprocess", workers = 15)
nCoV.integrated <- IntegrateData(anchorset = nCoV.anchors)
plan("sequential")
Merging dataset 4 into 2
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 7 into 3
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 6 into 3 7
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 8 into 5
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 1 into 2 4
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 9 into 3 7 6
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 12 into 10
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 13 into 2 4 1
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 5 8 into 3 7 6 9
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 2 4 1 13 into 10 12
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 11 into 10 12 2 4 1 13
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
Merging dataset 10 12 2 4 1 13 11 into 3 7 6 9 5 8
Extracting anchors for merged samples
Finding integration vectors
Finding integration vector weights
Integrating data
samples = read.delim2("./covid_meta.txt",header = TRUE, stringsAsFactors = FALSE,check.names = FALSE, sep = "\t")
# nCoV.integrated=sample.combined
sample_info = as.data.frame(colnames(nCoV.integrated))
colnames(sample_info) = c('ID')
rownames(sample_info) = sample_info$ID
sample_info$sample = nCoV.integrated@meta.data$orig.ident
sample_info = dplyr::left_join(sample_info,samples)
rownames(sample_info) = sample_info$ID
nCoV.integrated = AddMetaData(object = nCoV.integrated, metadata = sample_info)
Joining, by = "sample"
samples
| sample | sample_new | sample_new_old | group | disease | nCoV_mean | nFeature_RNA_low | nFeature_RNA_high | nCount_RNA_threshold | percent.mito |
|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <chr> | <chr> | <chr> | <chr> | <int> | <int> | <int> | <int> |
| C51 | HC1 | HC1 | HC | N | 0 | 200 | 6000 | 1000 | 10 |
| C52 | HC2 | HC2 | HC | N | 0 | 200 | 6000 | 1000 | 10 |
| C100 | HC3 | HC3 | HC | N | 0 | 200 | 6000 | 1000 | 10 |
| GSM3660650 | HC4 | HC4 | HC | N | 0 | 200 | 6000 | 1000 | 10 |
| C141 | M1 | O1 | M | Y | 0 | 200 | 6000 | 1000 | 10 |
| C142 | M2 | O2 | M | Y | 0 | 200 | 6000 | 1000 | 10 |
| C144 | M3 | O3 | M | Y | 0 | 200 | 6000 | 1000 | 10 |
| C145 | S1 | S1 | S | Y | 0.085901131 | 200 | 6000 | 1000 | 10 |
| C143 | S2 | C1 | S | Y | 0.007000048 | 200 | 6000 | 1000 | 10 |
| C146 | S3 | C2 | S | Y | 0.749939188 | 200 | 6000 | 1000 | 10 |
| C148 | S4 | C3 | S | Y | 0.00255102 | 200 | 6000 | 1000 | 10 |
| C149 | S5 | C4 | S | Y | 0.408822508 | 200 | 6000 | 1000 | 10 |
| C152 | S6 | C5 | S | Y | 0.096482152 | 200 | 6000 | 1000 | 10 |
DefaultAssay(nCoV.integrated) <- "integrated"
# Run the standard workflow for visualization and clustering
nCoV.integrated <- ScaleData(nCoV.integrated, verbose = FALSE)
nCoV.integrated <- RunPCA(nCoV.integrated, npcs = 30, verbose = FALSE)
nCoV.integrated <- RunUMAP(nCoV.integrated, reduction = "pca", dims = 1:30)
nCoV.integrated <- FindNeighbors(nCoV.integrated, reduction = "pca", dims = 1:30)
nCoV.integrated <- FindClusters(nCoV.integrated, resolution = 0.5)
Warning message:
“The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per session”
11:55:19 UMAP embedding parameters a = 0.9922 b = 1.112
11:55:19 Read 65706 rows and found 30 numeric columns
11:55:19 Using Annoy for neighbor search, n_neighbors = 30
11:55:19 Building Annoy index with metric = cosine, n_trees = 50
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
|
11:55:27 Writing NN index file to temp file /tmp/RtmpFpm1Oa/file18efb2a79ef27
11:55:27 Searching Annoy index using 1 thread, search_k = 3000
11:55:58 Annoy recall = 100%
11:55:58 Commencing smooth kNN distance calibration using 1 thread
11:56:03 Initializing from normalized Laplacian + noise
11:56:08 Commencing optimization for 200 epochs, with 3048454 positive edges
11:57:50 Optimization finished
Computing nearest neighbor graph
Computing SNN
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 65706
Number of edges: 2333410
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8994
Number of communities: 20
Elapsed time: 26 seconds
nCoV.integrated <- FindClusters(nCoV.integrated, resolution = 1.2)
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 65706
Number of edges: 2333410
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8600
Number of communities: 37
Elapsed time: 19 seconds
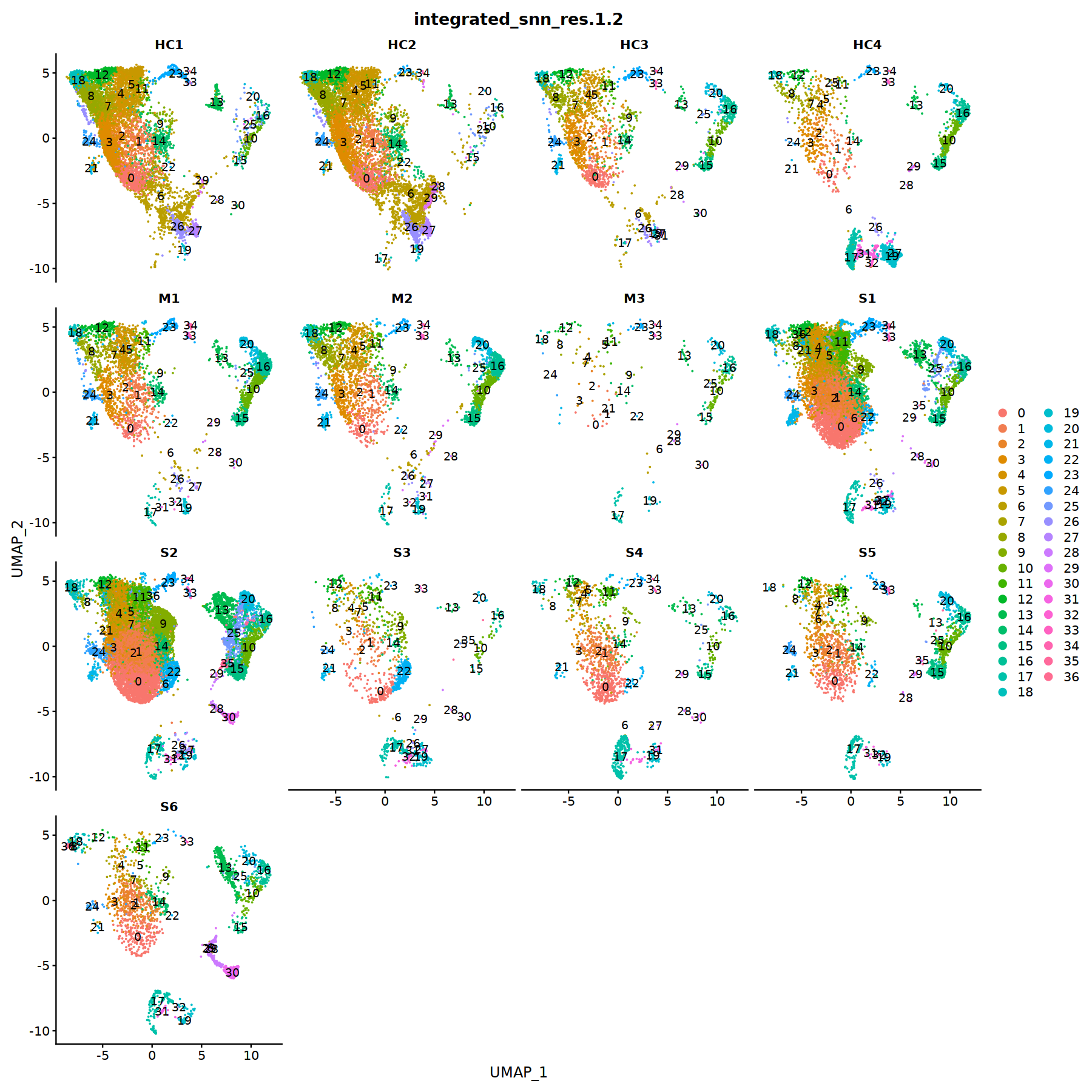
options(repr.plot.width=15, repr.plot.height=15)
DimPlot(object = nCoV.integrated, reduction = 'umap',label = TRUE,
group.by = 'integrated_snn_res.1.2',
split.by = 'sample_new', ncol = 4)
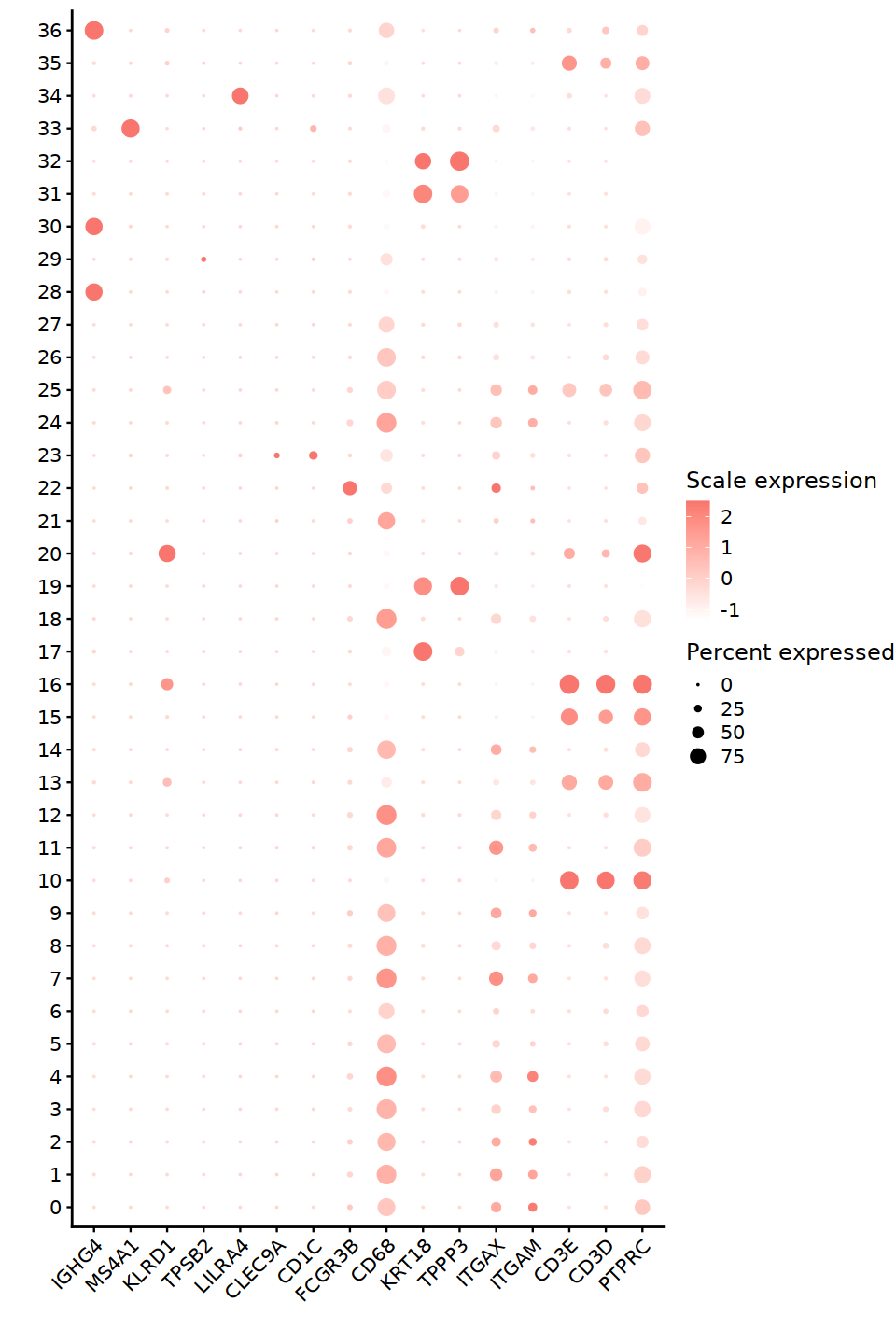
markers = c('PTPRC','CD3D','CD3E','ITGAM','ITGAX','TPPP3','KRT18','CD68','FCGR3B','CD1C','CLEC9A',
'LILRA4','TPSB2','KLRD1','MS4A1','IGHG4')
library(ggpubr)
DefaultAssay(nCoV.integrated)<-'RNA'
pp = DotPlot(nCoV.integrated, features = rev(markers),
cols = c('white','#F8766D'),dot.scale =5) + RotatedAxis()
pp = pp + theme(axis.text.x = element_text(size = 12),
axis.text.y = element_text(size = 12)) + labs(x='',y='') +
guides(color = guide_colorbar(title = 'Scale expression'),
size = guide_legend(title = 'Percent expressed')) +
theme(axis.line = element_line(size = 0.6))
o(8,12)
pp
o(12,12)
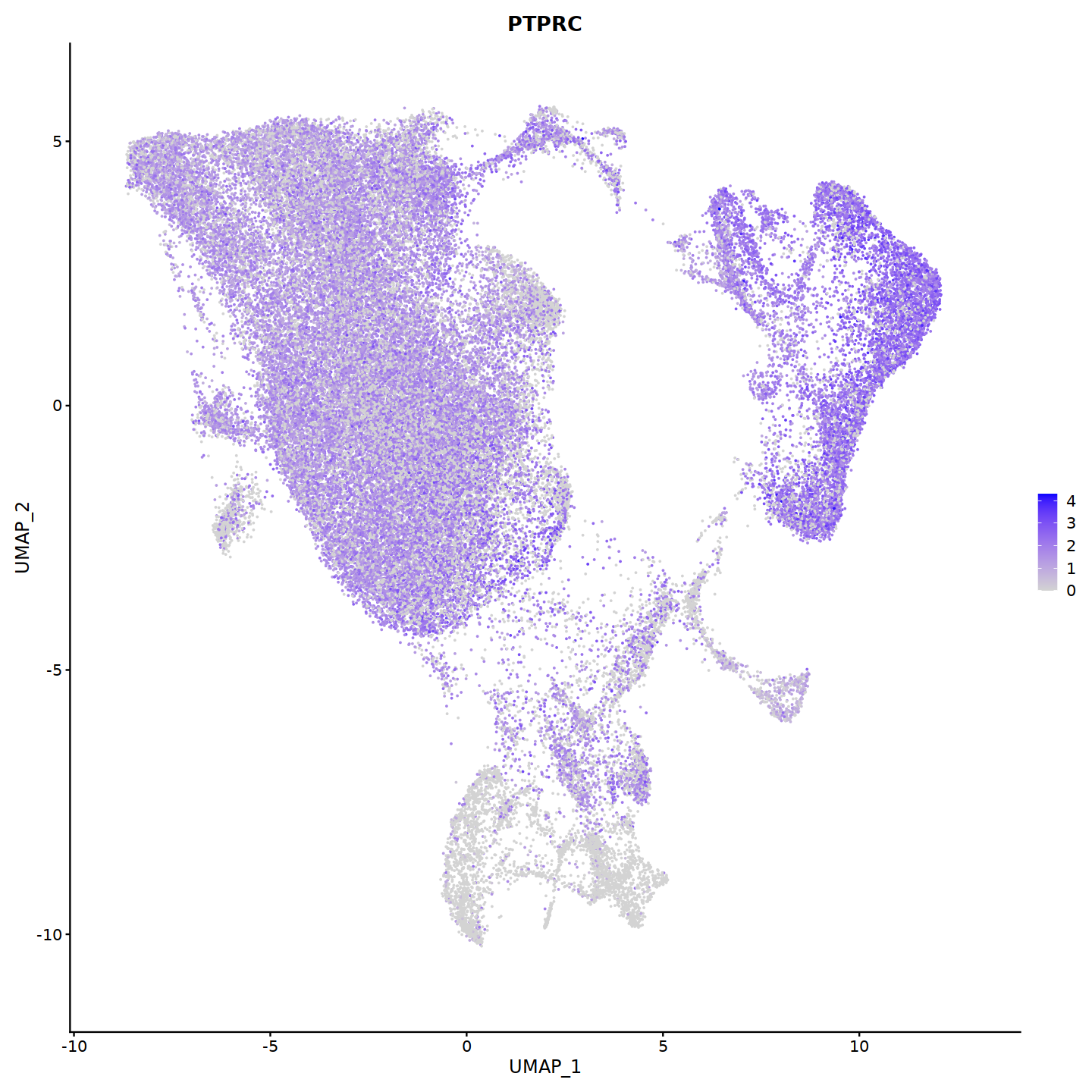
FeaturePlot(nCoV.integrated, features = 'PTPRC')
save(nCoV.integrated,
file = 'nCoV.integrated.rda',
compress = T, compression_level = 9)
nCoV.list = SplitObject(nCoV.integrated, split.by = 'sample_new')
dissociation.genes.hs<-c("ACTG1","ANKRD1","ARID5A","ATF3","ATF4","BAG3","BHLHE40","CCNL1","CCRN4L",
"CEBPB","CEBPD","CEBPG","CSRNP1","CXCL1","CYR61","DCN","DDX3XX","DDX5","DES","DNAJA1","DNAJB1",
"DNAJB4","DUSP1","DUSP8","EGR1","EGR2","EIF1","EIF5","ERF","ERRFI1","FAM132B","FOS","FOSB","FOSL2",
"GADD45A","GADD45G","BRD2","BTG1","BTG2","GCC1","GEM","H3F3B","HIPK3","HSP90AA1","HSP90AB1",
"HSPA1A","HSPA1B","HSPA5","HSPA8","HSPB1","HSPE1","HSPH1","ID3","IDI1","IER2","IER3","IER5",
"IFRD1","IL6","IRF1","IRF8","ITPKC","JUN","JUNB","JUND","KCNE4","KLF2","KLF4","KLF6","KLF9",
"LITAF","LMNA","MAFF","MAFK","MCL1","MIDN","MIR22HG","MT1","MT2","MYADM","MYC","MYD88","NCKAP5L",
"NCOA7","NFKBIA","NFKBIZ","NOP58","NPPC","NR4A1","ODC1","OSGIN1","OXNAD1","PCF11","PDE4B","PER1",
"PHLDA1","PNP","PNRC1","PPP1CC","PPP1R15A","PXDC1","RAP1B","RASSF1","RHOB","RHOH","RIPK1","SAT1X",
"SBNO2","SDC4","SERPINE1","SKIL","SLC10A6","SLC38A2","SLC41A1","SOCS3","SQSTM1","SRF","SRSF5",
"SRSF7","STAT3","TAGLN2","TIPARP","TNFAIP3","TNFAIP6","TPM3","TPPP3","TRA2A","TRA2B","TRIB1",
"TUBB4B","TUBB6","UBC","USP2","WAC","ZC3H12A","ZFAND5","ZFP36","ZFP36L1","ZFP36L2","ZYX")
# normalize and identify variable features for each dataset independently
nCoV.list <- lapply(X = nCoV.list, FUN = function(x) {
DefaultAssay(x)<-'RNA'
x <- NormalizeData(x, verbose = F)
x <- FindVariableFeatures(x, selection.method = "vst", nfeatures = 2000, verbose = F)
x <- ScaleData(x, features = rownames(x), verbose = F)
x <- RunPCA(object = x, features = VariableFeatures(x), npcs = 50, verbose = F)
x <- FindNeighbors(x, reduction = "pca", dims = 1:30, verbose = F)
x <- RunUMAP(object=x,reduction = "pca", dims = 1:30, verbose = F)
x <- FindClusters(object=x, resolution = c(0.7,0.9,1.2),verbose = F)
(x %>%
as.SingleCellExperiment %>%
cxds_bcds_hybrid)@colData[,c('cxds_score','bcds_score','hybrid_score')
] %>% as.data.frame -> scds.doublet.profiles
meta <- merge(x@meta.data, scds.doublet.profiles, by.x=0, by.y=0)
rownames(meta) <- meta$Row.names
meta$Row.names <- NULL
x@meta.data <- meta
gset <- dissociation.genes.hs
gset <- gset[gset %in% rownames(x)]
x[["percent.disso"]]<-PercentageFeatureSet(x, features = gset)
x
})
[12:39:39] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:40:41] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:43:14] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:43:28] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:45:22] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:45:50] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:46:20] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:46:54] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:47:35] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:49:23] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:50:57] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:51:44] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
[12:52:29] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
library(tictoc)
tic()
save(nCoV.list,
file = 'nCoV.list.rda',
compress = T, compression_level = 9)
toc()
2938.669 sec elapsed
i=1
nCoV.list[[i]][[]]
| orig.ident | nCount_RNA | nFeature_RNA | group | percent.mt | ID | sample | sample_new | sample_new_old | disease | ⋯ | integrated_snn_res.0.5 | seurat_clusters | integrated_snn_res.1.2 | RNA_snn_res.0.7 | RNA_snn_res.0.9 | RNA_snn_res.1.2 | cxds_score | bcds_score | hybrid_score | percent.disso | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <int> | <chr> | <dbl> | <chr> | <chr> | <chr> | <chr> | <chr> | ⋯ | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <dbl> | <dbl> | <dbl> | <dbl> | |
| AAACCTGAGATGTCGG-1_1 | C141 | 3731 | 1594 | M | 1.7421603 | AAACCTGAGATGTCGG-1_1 | C141 | M1 | O1 | Y | ⋯ | 7 | 2 | 10 | 0 | 3 | 2 | 7.385357e+03 | 0.0060111498 | 0.097049404 | 2.197802 |
| AAACCTGAGGCTCATT-1_1 | C141 | 33339 | 5273 | M | 1.6377216 | AAACCTGAGGCTCATT-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 1 | 5 | 2 | 1 | 1 | 6.435988e+04 | 0.1728955805 | 0.968124847 | 1.580731 |
| AAACCTGCAATCCGAT-1_1 | C141 | 5727 | 1789 | M | 0.2793784 | AAACCTGCAATCCGAT-1_1 | C141 | M1 | O1 | Y | ⋯ | 17 | 1 | 21 | 2 | 1 | 1 | 2.040284e-01 | 0.0012349773 | 0.001012450 | 1.606426 |
| AAACCTGCATGGTCAT-1_1 | C141 | 4396 | 2002 | M | 0.9326661 | AAACCTGCATGGTCAT-1_1 | C141 | M1 | O1 | Y | ⋯ | 7 | 5 | 15 | 4 | 5 | 5 | 1.060499e+04 | 0.0106119365 | 0.141438867 | 2.775250 |
| AAACCTGGTTTAGCTG-1_1 | C141 | 3219 | 1451 | M | 2.9201615 | AAACCTGGTTTAGCTG-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 2 | 16 | 0 | 3 | 2 | 5.606312e+03 | 0.0012850814 | 0.070335290 | 2.329916 |
| AAACCTGTCAATCACG-1_1 | C141 | 4002 | 1691 | M | 2.3488256 | AAACCTGTCAATCACG-1_1 | C141 | M1 | O1 | Y | ⋯ | 7 | 5 | 15 | 4 | 5 | 5 | 1.259333e+04 | 0.0446735024 | 0.200105882 | 2.948526 |
| AAACCTGTCCGAGCCA-1_1 | C141 | 26988 | 5497 | M | 2.1379873 | AAACCTGTCCGAGCCA-1_1 | C141 | M1 | O1 | Y | ⋯ | 6 | 6 | 8 | 2 | 1 | 6 | 7.821672e+03 | 0.0098174438 | 0.106251123 | 2.141693 |
| AAACCTGTCCTCCTAG-1_1 | C141 | 1118 | 559 | M | 1.2522361 | AAACCTGTCCTCCTAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 17 | 10 | 21 | 6 | 7 | 10 | 7.471562e+02 | 0.0010683149 | 0.010075433 | 1.073345 |
| AAACGGGAGAACTCGG-1_1 | C141 | 4223 | 1768 | M | 1.8233483 | AAACGGGAGAACTCGG-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 2 | 16 | 0 | 3 | 2 | 8.899078e+03 | 0.0328886285 | 0.142659920 | 1.799668 |
| AAACGGGAGTCAAGCG-1_1 | C141 | 19045 | 3571 | M | 1.8955106 | AAACGGGAGTCAAGCG-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 0 | 3 | 1 | 0 | 0 | 2.099826e+03 | 0.0076494459 | 0.033378011 | 1.575217 |
| AAACGGGCACTCTGTC-1_1 | C141 | 28604 | 4594 | M | 2.8527479 | AAACGGGCACTCTGTC-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 0 | 1 | 1 | 0 | 0 | 6.093326e+04 | 0.0338052474 | 0.786545420 | 1.408894 |
| AAACGGGCAGCGTAAG-1_1 | C141 | 25832 | 4283 | M | 2.4698049 | AAACGGGCAGCGTAAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 1.060602e+04 | 0.0086387368 | 0.139476315 | 1.238774 |
| AAACGGGGTGGAAAGA-1_1 | C141 | 2969 | 1488 | M | 1.9198383 | AAACGGGGTGGAAAGA-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 3 | 16 | 0 | 4 | 3 | 6.573174e+03 | 0.0070918938 | 0.088095429 | 2.222971 |
| AAACGGGTCGTTACAG-1_1 | C141 | 32695 | 4949 | M | 1.7831473 | AAACGGGTCGTTACAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 8 | 1 | 3 | 2 | 8 | 6.494869e+03 | 0.2911204398 | 0.371457852 | 1.578223 |
| AAACGGGTCTTCATGT-1_1 | C141 | 11766 | 3014 | M | 1.6403196 | AAACGGGTCTTCATGT-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 8 | 1 | 3 | 2 | 8 | 1.831672e+03 | 0.0049657403 | 0.027377975 | 1.946286 |
| AAACGGGTCTTGCAAG-1_1 | C141 | 14341 | 3900 | M | 0.8018967 | AAACGGGTCTTGCAAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 6 | 3 | 2 | 1 | 6 | 1.167642e+04 | 0.0189507920 | 0.163025886 | 1.506171 |
| AAAGATGAGTAAGTAC-1_1 | C141 | 25515 | 4162 | M | 4.9970606 | AAAGATGAGTAAGTAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 5.938664e+04 | 0.0065685916 | 0.740168788 | 1.262003 |
| AAAGATGAGTGGTCCC-1_1 | C141 | 1050 | 706 | M | 3.9047619 | AAAGATGAGTGGTCCC-1_1 | C141 | M1 | O1 | Y | ⋯ | 7 | 2 | 15 | 0 | 3 | 2 | 1.802625e+02 | 0.0008281182 | 0.002830077 | 3.142857 |
| AAAGATGCAATGGAGC-1_1 | C141 | 30605 | 4902 | M | 1.1370691 | AAAGATGCAATGGAGC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 1.266751e+04 | 0.0380518027 | 0.194393802 | 1.421336 |
| AAAGATGCACTCTGTC-1_1 | C141 | 16594 | 3972 | M | 0.3977341 | AAAGATGCACTCTGTC-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 6 | 3 | 2 | 1 | 6 | 5.887888e+03 | 0.0187465176 | 0.091294587 | 2.115222 |
| AAAGATGGTCTAGTCA-1_1 | C141 | 31509 | 5159 | M | 1.2631312 | AAAGATGGTCTAGTCA-1_1 | C141 | M1 | O1 | Y | ⋯ | 6 | 6 | 8 | 2 | 1 | 6 | 7.689386e+03 | 0.0246478207 | 0.119462630 | 1.837570 |
| AAAGATGTCACTCTTA-1_1 | C141 | 26748 | 4709 | M | 3.4058621 | AAAGATGTCACTCTTA-1_1 | C141 | M1 | O1 | Y | ⋯ | 3 | 0 | 1 | 1 | 0 | 0 | 6.605723e+04 | 0.0029935390 | 0.819016078 | 1.936593 |
| AAAGATGTCAGGTTCA-1_1 | C141 | 4244 | 1775 | M | 2.5212064 | AAAGATGTCAGGTTCA-1_1 | C141 | M1 | O1 | Y | ⋯ | 13 | 3 | 20 | 0 | 4 | 3 | 8.488594e+03 | 0.0047744564 | 0.109443696 | 4.853911 |
| AAAGATGTCGGATGGA-1_1 | C141 | 3896 | 1635 | M | 1.4887064 | AAAGATGTCGGATGGA-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 2 | 10 | 0 | 3 | 2 | 7.223007e+03 | 0.0080970153 | 0.097131373 | 2.977413 |
| AAAGATGTCTCCAACC-1_1 | C141 | 28573 | 4622 | M | 1.3299269 | AAAGATGTCTCCAACC-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 0 | 1 | 1 | 2 | 0 | 5.844056e+03 | 0.0256461129 | 0.097659894 | 2.190880 |
| AAAGCAAAGTGAAGAG-1_1 | C141 | 21282 | 5981 | M | 6.7333897 | AAAGCAAAGTGAAGAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 10 | 16 | 19 | 11 | 14 | 16 | 3.519066e+03 | 0.1239518374 | 0.167340883 | 2.678320 |
| AAAGCAACAGCGTCCA-1_1 | C141 | 2425 | 1324 | M | 0.9484536 | AAAGCAACAGCGTCCA-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 2 | 10 | 0 | 3 | 2 | 7.927301e+03 | 0.0070140962 | 0.104750017 | 2.268041 |
| AAAGCAACAGTGAGTG-1_1 | C141 | 7455 | 2468 | M | 3.5144199 | AAAGCAACAGTGAGTG-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 19 | 33 | 6 | 16 | 19 | 1.302528e+04 | 0.0159199145 | 0.176659165 | 1.703555 |
| AAAGCAAGTCATATCG-1_1 | C141 | 8676 | 3094 | M | 2.9276164 | AAAGCAAGTCATATCG-1_1 | C141 | M1 | O1 | Y | ⋯ | 3 | 7 | 0 | 5 | 6 | 7 | 2.711236e+03 | 0.0083096186 | 0.041593854 | 2.109267 |
| AAAGCAAGTCCAACTA-1_1 | C141 | 11768 | 3144 | M | 1.5550646 | AAAGCAAGTCCAACTA-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 1 | 8 | 2 | 1 | 1 | 5.152539e+03 | 0.0295560323 | 0.093029131 | 1.784500 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| TTTATGCCAATTGCTG-1_1 | C141 | 29946 | 4513 | M | 4.9255326 | TTTATGCCAATTGCTG-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 14050.3761 | 0.214436278 | 0.38805301 | 1.075269 |
| TTTATGCCACAACTGT-1_1 | C141 | 21759 | 4880 | M | 1.2362700 | TTTATGCCACAACTGT-1_1 | C141 | M1 | O1 | Y | ⋯ | 6 | 6 | 8 | 2 | 1 | 6 | 11375.0096 | 0.064012140 | 0.20441067 | 1.971598 |
| TTTATGCCATGCGCAC-1_1 | C141 | 12507 | 3170 | M | 0.6236508 | TTTATGCCATGCGCAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 4 | 12 | 3 | 2 | 4 | 46454.8787 | 0.177405417 | 0.75139375 | 1.918925 |
| TTTATGCTCCTAGAAC-1_1 | C141 | 18715 | 4258 | M | 3.9593909 | TTTATGCTCCTAGAAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 16 | 13 | 24 | 2 | 11 | 13 | 715.9992 | 0.009067236 | 0.01769785 | 1.768635 |
| TTTCCTCAGAGTTGGC-1_1 | C141 | 4631 | 1927 | M | 2.3752969 | TTTCCTCAGAGTTGGC-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 3 | 16 | 0 | 4 | 3 | 11732.5469 | 0.009758063 | 0.15451697 | 2.742388 |
| TTTCCTCAGCGTTGCC-1_1 | C141 | 21023 | 4499 | M | 3.0490415 | TTTCCTCAGCGTTGCC-1_1 | C141 | M1 | O1 | Y | ⋯ | 16 | 0 | 24 | 1 | 0 | 0 | 11583.0294 | 0.007007261 | 0.14991571 | 1.393712 |
| TTTCCTCAGGAATCGC-1_1 | C141 | 20205 | 4199 | M | 6.2261816 | TTTCCTCAGGAATCGC-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 7 | 4 | 5 | 6 | 7 | 53633.1626 | 0.423103780 | 1.08605238 | 2.395447 |
| TTTCCTCCACACTGCG-1_1 | C141 | 32457 | 4950 | M | 2.4894476 | TTTCCTCCACACTGCG-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 4 | 1 | 3 | 2 | 4 | 5575.4627 | 0.054871671 | 0.12359756 | 2.412423 |
| TTTCCTCCAGCGAACA-1_1 | C141 | 24540 | 4654 | M | 3.2192339 | TTTCCTCCAGCGAACA-1_1 | C141 | M1 | O1 | Y | ⋯ | 3 | 1 | 0 | 2 | 1 | 1 | 56334.8208 | 0.035879195 | 0.73180025 | 1.894866 |
| TTTCCTCGTGAGTATA-1_1 | C141 | 32663 | 5670 | M | 0.8725469 | TTTCCTCGTGAGTATA-1_1 | C141 | M1 | O1 | Y | ⋯ | 6 | 1 | 18 | 2 | 1 | 1 | 2924.4883 | 0.151121661 | 0.18719256 | 1.898172 |
| TTTCCTCTCCTGTACC-1_1 | C141 | 21555 | 4213 | M | 1.2479703 | TTTCCTCTCCTGTACC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 7 | 11 | 5 | 6 | 7 | 47586.2856 | 0.009095574 | 0.59688566 | 2.635119 |
| TTTCCTCTCGCGCCAA-1_1 | C141 | 4338 | 1908 | M | 1.2448133 | TTTCCTCTCGCGCCAA-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 5 | 10 | 4 | 5 | 5 | 8836.6182 | 0.007442442 | 0.11641493 | 2.558783 |
| TTTGCGCAGGGCTTCC-1_1 | C141 | 8499 | 2905 | M | 2.3532180 | TTTGCGCAGGGCTTCC-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 2 | 16 | 0 | 3 | 2 | 17940.0904 | 0.159516767 | 0.38113902 | 1.753147 |
| TTTGCGCCAATTGCTG-1_1 | C141 | 12099 | 2462 | M | 1.8431275 | TTTGCGCCAATTGCTG-1_1 | C141 | M1 | O1 | Y | ⋯ | 3 | 9 | 1 | 7 | 8 | 9 | 31147.6082 | 0.026614668 | 0.41129646 | 2.694438 |
| TTTGCGCGTGCTCTTC-1_1 | C141 | 38216 | 4877 | M | 1.1435001 | TTTGCGCGTGCTCTTC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 4 | 12 | 3 | 2 | 4 | 8070.5393 | 0.153765932 | 0.25342755 | 1.452271 |
| TTTGCGCTCACCCTCA-1_1 | C141 | 20602 | 3904 | M | 4.7082807 | TTTGCGCTCACCCTCA-1_1 | C141 | M1 | O1 | Y | ⋯ | 8 | 1 | 14 | 2 | 1 | 1 | 7696.1792 | 0.019599268 | 0.11449266 | 1.970682 |
| TTTGCGCTCCGTAGGC-1_1 | C141 | 23267 | 3648 | M | 1.8781966 | TTTGCGCTCCGTAGGC-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 9 | 23 | 7 | 8 | 9 | 30037.0625 | 0.006686029 | 0.37762406 | 3.752095 |
| TTTGCGCTCTACTTAC-1_1 | C141 | 5113 | 1880 | M | 3.2857422 | TTTGCGCTCTACTTAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 19 | 33 | 6 | 16 | 19 | 10664.8692 | 0.005723372 | 0.13728508 | 1.642871 |
| TTTGCGCTCTTGCAAG-1_1 | C141 | 23433 | 4050 | M | 1.2717108 | TTTGCGCTCTTGCAAG-1_1 | C141 | M1 | O1 | Y | ⋯ | 6 | 0 | 12 | 1 | 0 | 0 | 6955.2617 | 0.052789316 | 0.13856268 | 1.412538 |
| TTTGCGCTCTTTACGT-1_1 | C141 | 28584 | 4495 | M | 3.9707529 | TTTGCGCTCTTTACGT-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 0 | 1 | 1 | 2 | 0 | 64083.9704 | 0.035655819 | 0.82733012 | 1.668766 |
| TTTGGTTAGCACGCCT-1_1 | C141 | 3782 | 1669 | M | 1.5071391 | TTTGGTTAGCACGCCT-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 3 | 10 | 0 | 4 | 3 | 7415.4439 | 0.010300782 | 0.10171536 | 2.564781 |
| TTTGGTTAGTGGTAAT-1_1 | C141 | 10560 | 3098 | M | 1.3731061 | TTTGGTTAGTGGTAAT-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 17 | 34 | 12 | 15 | 17 | 21858.4122 | 0.178794533 | 0.44885456 | 1.903409 |
| TTTGGTTAGTTGTAGA-1_1 | C141 | 32516 | 4967 | M | 2.0943535 | TTTGGTTAGTTGTAGA-1_1 | C141 | M1 | O1 | Y | ⋯ | 0 | 0 | 1 | 1 | 2 | 0 | 7371.8300 | 0.002813193 | 0.09368090 | 1.556157 |
| TTTGGTTCATACTACG-1_1 | C141 | 21735 | 4314 | M | 2.6224983 | TTTGGTTCATACTACG-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 62425.0614 | 0.046187248 | 0.81737418 | 1.352657 |
| TTTGTCAAGATTACCC-1_1 | C141 | 23375 | 4483 | M | 1.0695187 | TTTGTCAAGATTACCC-1_1 | C141 | M1 | O1 | Y | ⋯ | 2 | 1 | 8 | 2 | 1 | 1 | 1660.2334 | 0.118737273 | 0.13915185 | 1.112299 |
| TTTGTCAAGTGGTAAT-1_1 | C141 | 4123 | 1831 | M | 2.5466893 | TTTGTCAAGTGGTAAT-1_1 | C141 | M1 | O1 | Y | ⋯ | 5 | 3 | 16 | 0 | 4 | 3 | 1208.0658 | 0.079246655 | 0.09403204 | 2.110114 |
| TTTGTCACAGAAGCAC-1_1 | C141 | 8929 | 2971 | M | 2.5646769 | TTTGTCACAGAAGCAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 19 | 33 | 6 | 16 | 19 | 21351.9277 | 0.012140534 | 0.27576523 | 1.377534 |
| TTTGTCATCAACCAAC-1_1 | C141 | 4335 | 1939 | M | 2.4913495 | TTTGTCATCAACCAAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 7 | 2 | 15 | 0 | 3 | 2 | 9693.4556 | 0.005780285 | 0.12533864 | 2.491349 |
| TTTGTCATCCAAACAC-1_1 | C141 | 13918 | 3531 | M | 3.2332232 | TTTGTCATCCAAACAC-1_1 | C141 | M1 | O1 | Y | ⋯ | 12 | 9 | 23 | 7 | 8 | 9 | 26399.6501 | 0.046769656 | 0.37280402 | 1.501653 |
| TTTGTCATCGCGTTTC-1_1 | C141 | 25531 | 4537 | M | 2.2834985 | TTTGTCATCGCGTTTC-1_1 | C141 | M1 | O1 | Y | ⋯ | 1 | 0 | 5 | 1 | 0 | 0 | 51298.6899 | 0.006475167 | 0.64013533 | 1.335631 |
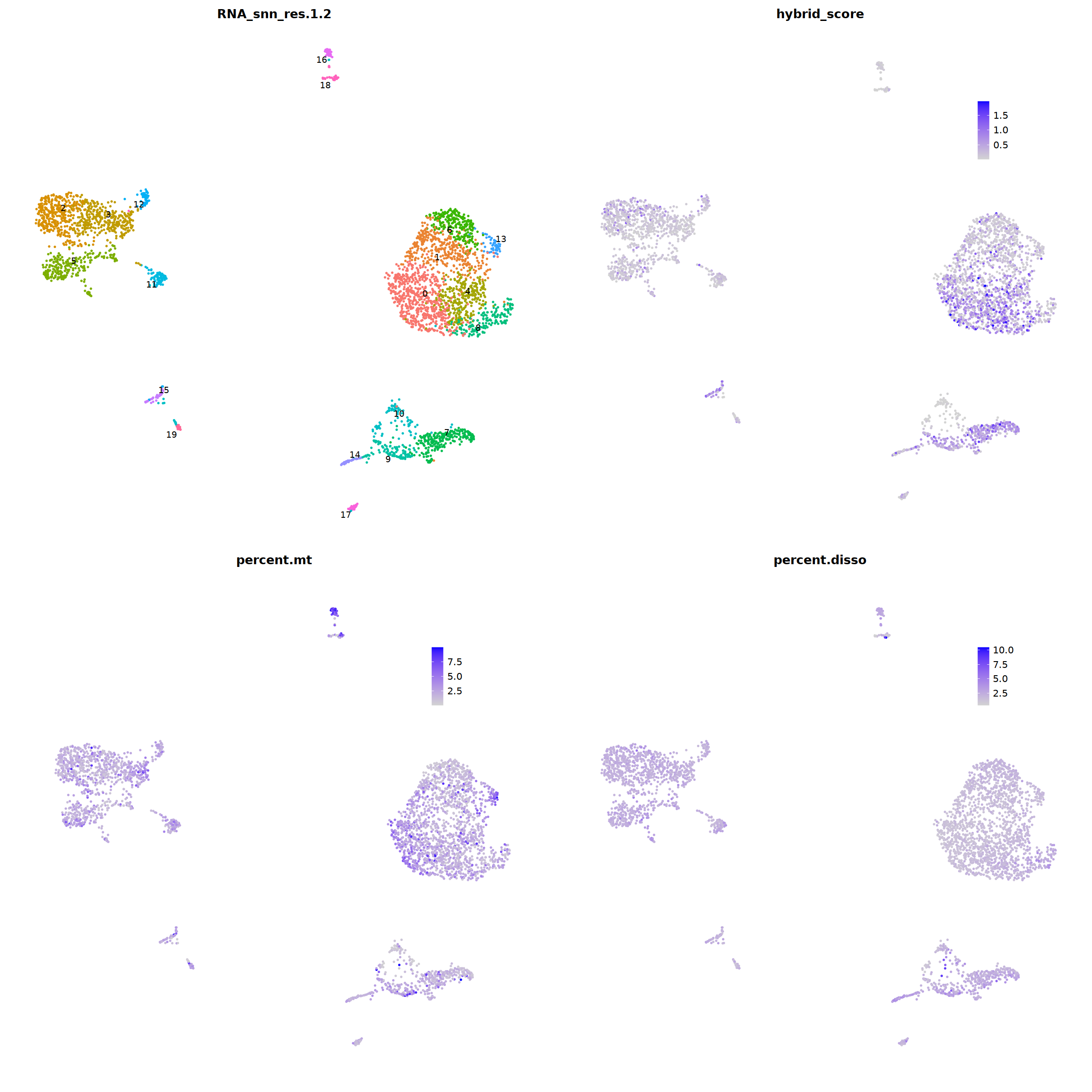
Quality control¶
i=1
o(20,20)
ggarrange(
DimPlot(nCoV.list[[i]], label=T,repel=T, group.by="RNA_snn_res.1.2")&NoLegend()&
theme(axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'hybrid_score')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.mt')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
FeaturePlot(nCoV.list[[i]], features = 'percent.disso')&
theme(legend.position=c(0.8,0.8),
axis.line = element_blank(),
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks=element_blank()
),
ncol=2,nrow=2
)
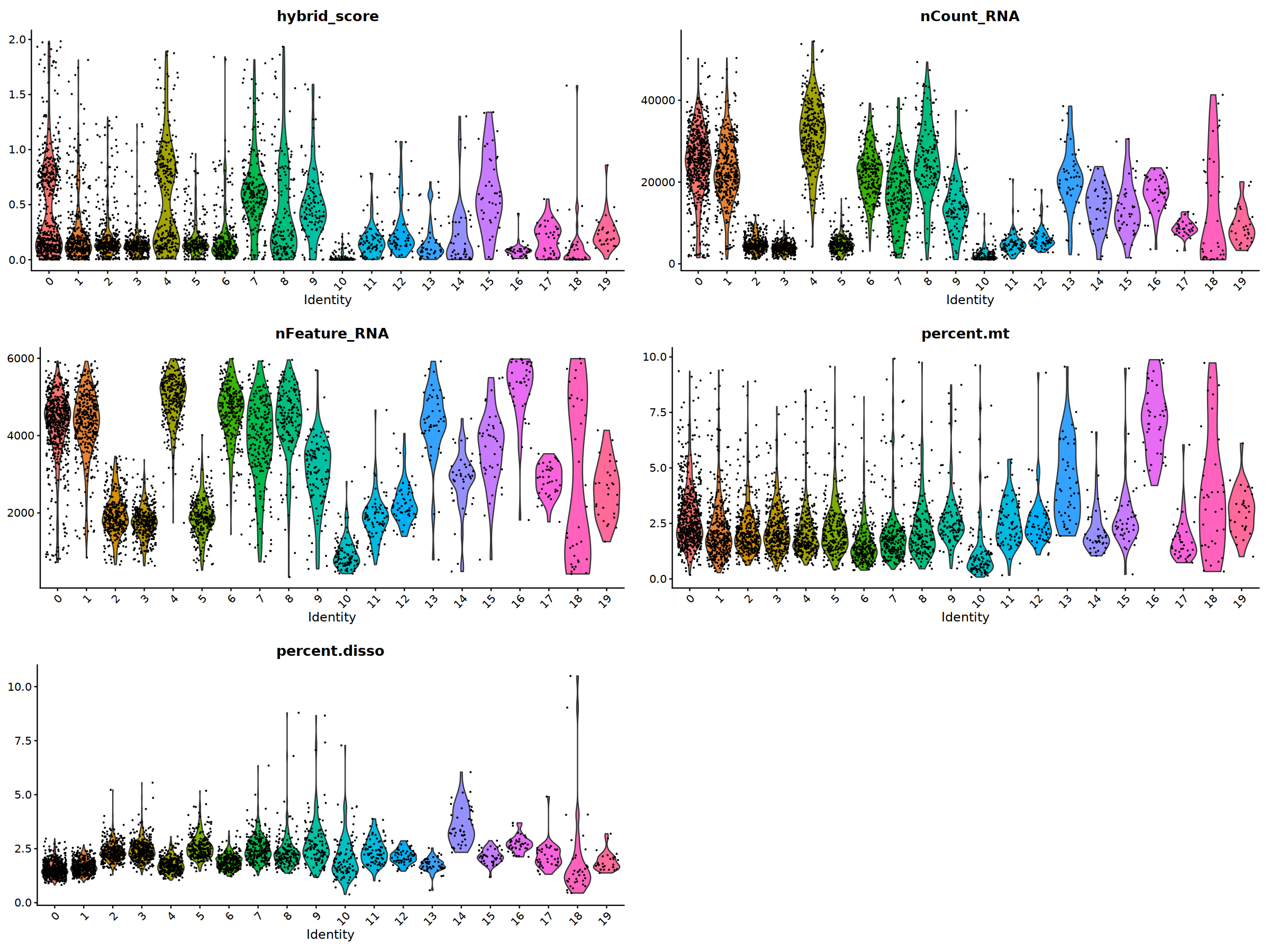
o(20,15)
ggarrange(
VlnPlot(nCoV.list[[i]], features = 'hybrid_score')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nCount_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'nFeature_RNA')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.mt')&NoLegend(),
VlnPlot(nCoV.list[[i]], features = 'percent.disso')&NoLegend(),
ncol=2, nrow=3
)